Figure 6.
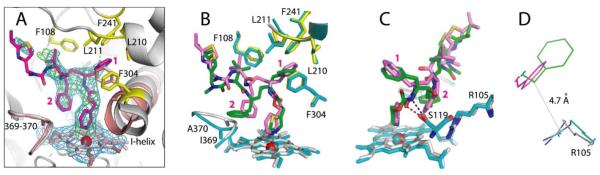
Crystal structure of the CYP3A4–3 complex. (A) Compound 3 (in magenta) binds to the heme via the pyridine nitrogen. Most of the ligand–protein interactions are provided by the phenyl side groups, labeled as (1) and (2). Phenyl-1 is imbedded into a hydrophobic pocket lined with Phe108, Leu210, Leu211, Phe241, and Phe304 (shown in yellow sticks). Steric clashing between Phenyl-1 and Phe304, which is also observed in the CYP3A4–ritonair complex, leads to the I-helix displacement. Phenyl-2 of 3, however, imposes no steric hindrance on the 369–370 peptide and, as a result, there is no heme shift. For comparison, the heme, 369–370 peptide, and I-helix of the superimposed ligand-free 1TQN structure are shown in pink. 2Fo – Fc and Fo – Fc electron density maps contoured at 1σ and 3σ are displayed in blue and green, respectively. (B,C) Relative orientation of 3 (magenta) and ritonavir (green). The heme, 369–370 peptide, and residues lining the hydrophobic pocket in the ritonavir-bound 3NXU structure are colored in cyan. Owing to higher flexibility, 3 binds in a conformation that eliminates clashing between Phenyl-2 and Ile369-Ala370 (B). Furthermore, Phenyl-2 orients near parallel to the pyridine ring and the guanidinium group of Arg105. This promotes π-stacking and cation–π interactions, stabilizing the complex. A peptide flip in 3 allows formation of a stronger hydrogen bond with Ser119 via the amide nitrogen (depicted as a blue dashed line in (C)). In contrast, ritonavir is H-bonded to Ser119 via the carbonyl oxygen (red dashed line). Such conformational differences could increase the binding affinity and inhibitory potency of compound 3. (D) A closer view at the Phe-2–Arg105 interaction site demonstrates that only the phenyl ring of 3 (magenta) can establish cation–π interactions with the Arg105 guanidinium group.
