Figure 1.
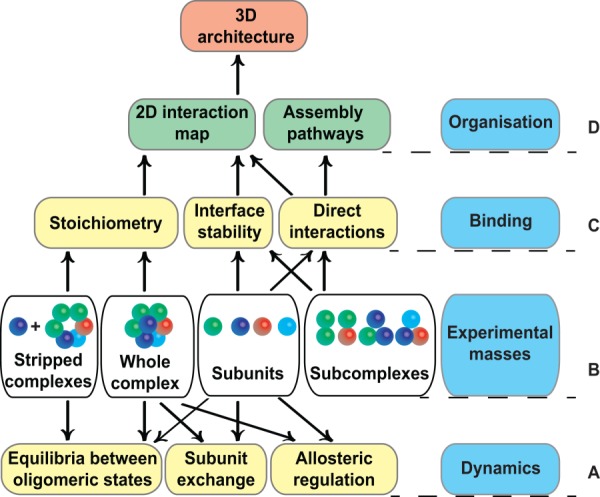
Native MS provides diverse information about macromolecular complexes. Primary data concerning the masses of the holo-complex, subcomplexes, and individual subunits (row B) provide access to a wealth of secondary information, such as binding interactions and dynamic properties (rows A, C), which in turn allows for higher inferences to be made concerning the organization of the complex (row D). Rows B-C: The experimental masses of individual subunits of a complex are determined following chromatographic separation under denaturing conditions.157 The use of MS conditions optimised for preserving noncovalent interactions (i.e., native MS) allows the mass of the intact complex to be measured. The combined subunit and holo-complex data reveal the stoichiometry of subunits, which can be further confirmed by a series of tandem MS spectra (i.e., “gas-phase dissociation” to generate stripped complexes32). The use of perturbing conditions or destabilizing agents (“in-solution dissociation”) allows one to generate overlapping subcomplexes (dimers, trimers, etc.), whose composition reveals direct interactions between subunits47 and allows interface stability to be assessed.13 Row D: The above data allow one to determine the two dimensional (2D) interaction network of subunits within a protein complex and, when combined with structural data (EM or SAXS envelopes), to deduce the 3D architecture of a macromolecular assembly (e.g., Refs.90 and91). In addition, individual subunits (or subcomplexes) can be mixed in solution and a mass shift can be detected if a (sub-)complex is generated (e.g., Ref.158), allowing the assembly pathway of a macromolecular complex to be identified. Row A: Native MS can provide information regarding the dynamic behavior of complexes. First, it can reveal the presence of different oligomeric states and monitor changes in equilibrium induced by different solution pH values and concentrations.5 Second, the subunit composition of intact complexes can be varied and monitored as a function of time by incubating light and heavy isoforms of a protein (e.g., labeled with 13C and 15N). The kinetics of subunit exchange can reveal distinct pathways for wild-type and mutant proteins (e.g., involved in amyloidosis,159 or assessing the effect of substrates and products.57) Third, native MS can relatively quantify the populations of macromolecular complexes containing different numbers of bound molecules, providing information about allostery.27
