Figure 6.
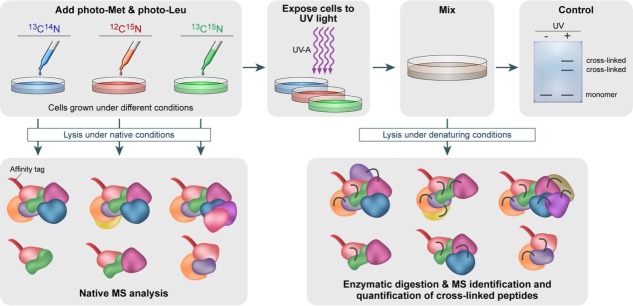
Schematic representation of an in vivo strategy for identifying protein–protein interactions. Cells are grown in media that are devoid of leucine and methionine. Photoreactive leucine and methionine analogues substitute the naturally occurring amino acids. To determine how multiple growth conditions influence the composition of a protein complex, cell cultures treated under different conditions will be cultured in different isotope-labeled media, containing the combinations of 13C- and 15N-labeling. Chemical crosslinking is then induced by UV-A illumination, forming a covalent bond with nearby protein side chains. Protein–protein interactions are identified by combining denaturing and native lysis conditions with pull-down experiments, using an affinity tag. Pull-down experiments under native conditions, before the formation of crosslinks, will enable to define subunit stoichiometries and characterization of all interacting proteins within the complex, using native MS. This step can also be performed in the absence of isotope labeling. Illuminating the sample with UV-A light will produce crosslinks capable of capturing even transient and weak interactions. These will be identified after pull-down experiments under denaturing conditions and analysis by both native MS and peptide MS profiling. The former provides a direct identification of the protein–protein interaction sites. MS/MS analysis of crosslinked products allows quantifying altered protein complex formation upon altered biological conditions based on 13C- and 15N-labeling.
