Fig. S7.
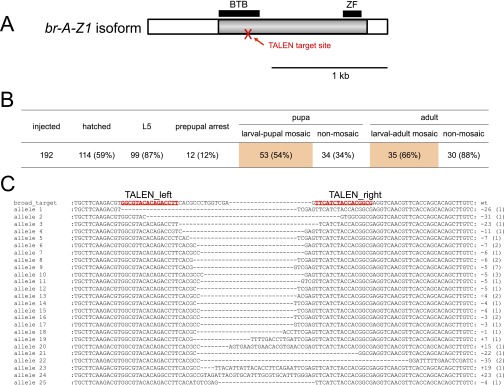
Mosaic analysis of broad. (A) Schematic representation of broad cDNA and the TALEN target site. Note that only the A-Z1 isoform is shown here, but there are at least 14 splice variants of broad in Bombyx (22). The target site was designed in the Bric-a-brac-Tramtrack-Broad (BTB) domain because this domain is present in all the broad isoforms that have been identified. Gray and white bars indicate the coding regions and the UTRs, respectively. The TALEN target site is indicated by a red arrow. ZF, zinc finger domain. (Scale bar, 1 kb.) (B) Summary of a mosaic analysis of broad. In this experiment, the dose of TALEN mRNAs was reduced to 4 ng/µL, 100-fold lower than the dose used in standard experiments (Materials and Methods in the main text). Among 114 hatched larvae, 53 became larval–pupal mosaics, and 35 became larval–adult mosaics (Fig. 7 in the main text). (C) Mutations induced by TALENs that targeted broad. TALEN mRNAs (400 ng/µL; note that this is a standard dose) were injected into embryos, and the hatched larvae (G0) were reared. Most of the larvae were dead by the prepupal stage, and none became pupae. gDNA was extracted from a larva that died at the prepupal stage and was used as a template for genomic PCR. PCR products were cloned into a cloning vector, and 48 clones were selected randomly and sequenced. In total, 38 clones were sequenced successfully, and all had mutations (100%). We recovered 25 mutant alleles with short insertions and deletions, which ranged from 23-bp insertions to 35-bp deletions; their nucleotide sequences are shown in the figure. The numbers in parentheses indicate the number of clones. The TALEN-binding sites are indicated by red letters.
