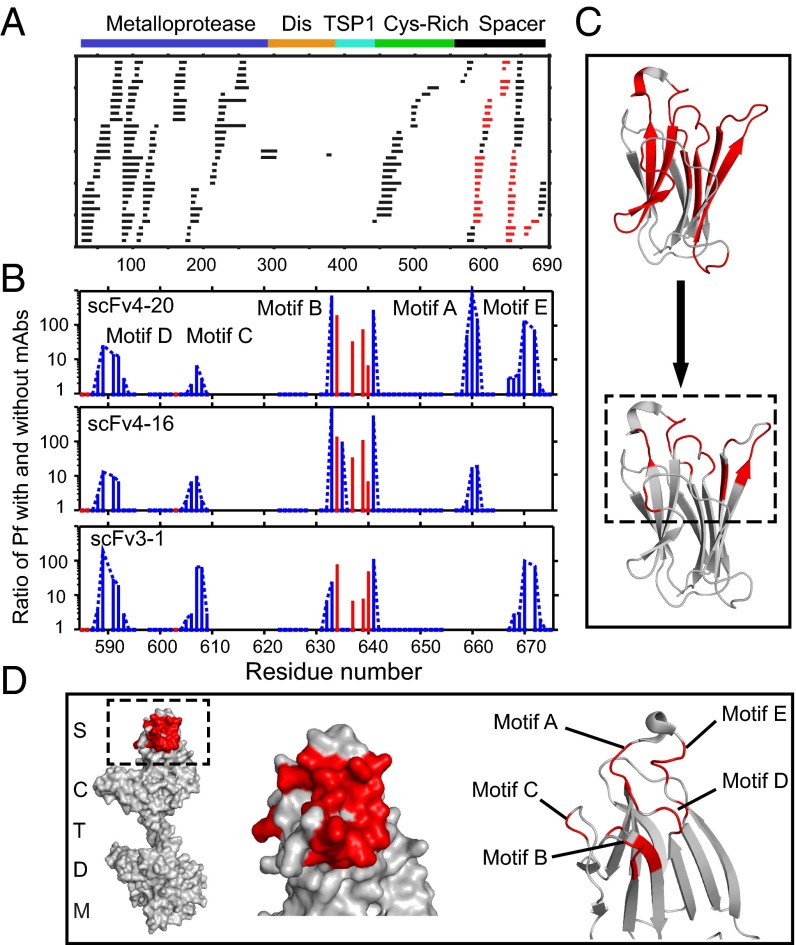
Fig. 3.
Antibody-binding sites in ADAMTS13 revealed by HX MS. (A) Acid protease digestion of human MDTCS fragment produced 208 unique overlapping peptides with 70 derived from the spacer domain and 113 from M. Black peptides show no difference in HX rate with and without scFv bound; red peptides show slowed HX rate upon scFv binding. (B) The ratio of additional protection factor (Pf) with scFv bound. Resolved single residues are in red; switchable residues in blue are connected by dashed lines. Some otherwise-protected regions missing in motifs A and E are ambiguous due to fewer peptides recovered and not necessarily to the absence of antigen–antibody interaction. (C) Representation of the binding epitope (red) at course peptide resolution (Top) and at refined residue resolution (Bottom). (D) Representations of the binding epitope (red) in the spacer domain of ADAMTS13. These results specify the binding epitope in terms of HX slowing in the five-loop region but do not necessarily indicate that the particular residues that show slowed exchange engage in specific interprotein interactions, for reasons described in Discussion.