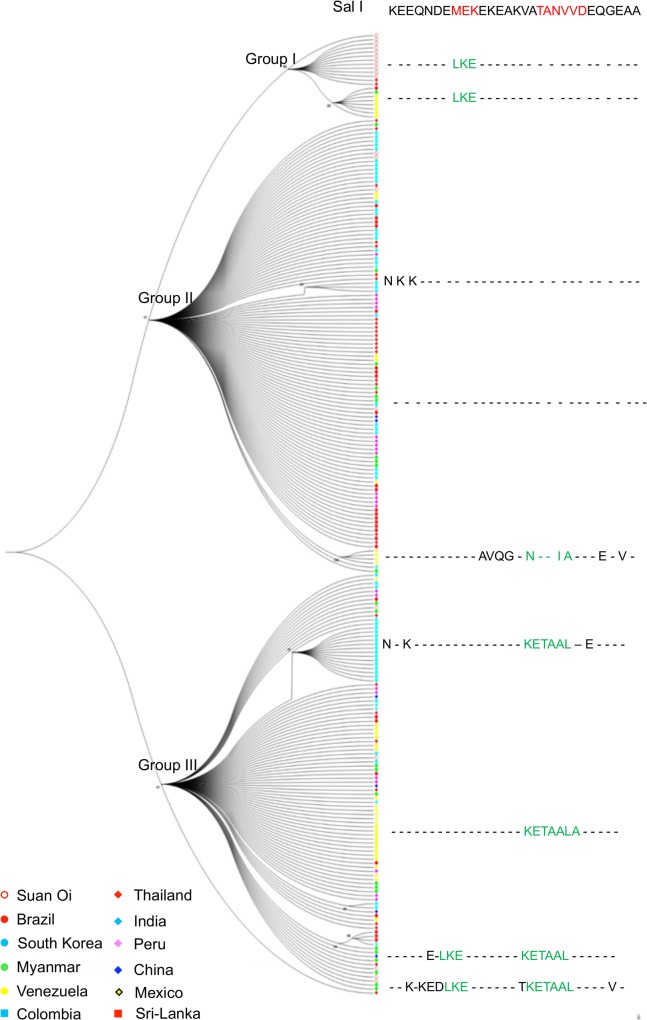
Fig 4. Maximum likelihood phylogeny of PvMSP3α block II DNA sequences.
An unrooted phylogeny of the 237 PvMSP3α block II sequences was inferred with maximum likelihood using Tamura and Nei’s model of nucleotide substitution implemented in MEGA6. Bootstrapping was performed with 500 replicates and tree was condensed using 75% bootstrap as a threshold. The label of each sequence is color coded corresponding to the country of origin. Three major clusters were identified as group I, II and III and several sub-clusters were observed within each group. Consensus protein sequences generated for each cluster/sub-cluster using 29 parsimony informative amino acid changes have been shown. Salvador I sequence has been shown as a reference. Amino acids highlighted in red are indicating wild alleles of structural motif I and II while amino acids in green are the mutated alleles. Dashes '-' are representing amino acids that are similar in all the clusters.