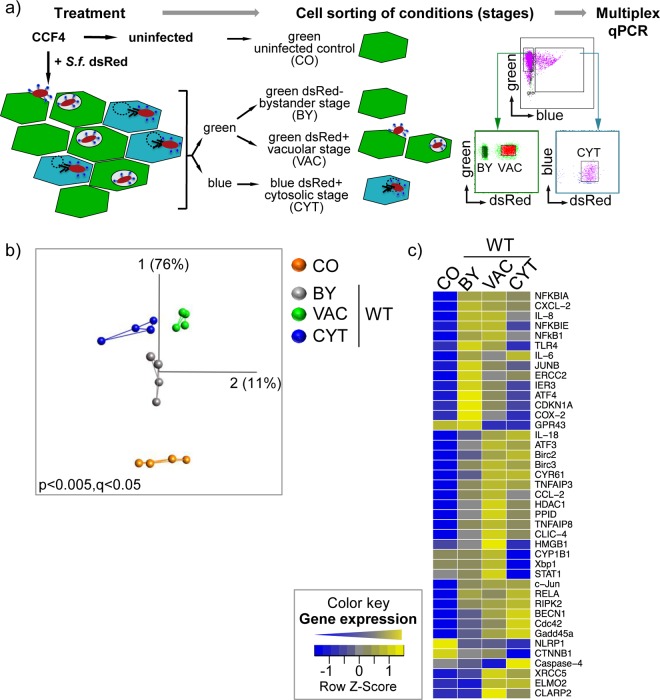
FIG 1.
Identification of localization-dependent transcriptional signatures during Shigella infection, combining fluorescence reporters and microfluidics. (a) Workflow for treatment, cell sorting, and subsequent transcriptional analysis of uninfected (CO) cells or cells at distinct stages of dsRed-expressing Shigella (+S.f. dsRed) infection (BY, VAC, and CYT). The conditions were distinguished using the CCF4/β-lactamase FRET approach and dsRed-fluorescent bacteria and were sorted by FACS according to their specific fluorescent properties, as indicated. The sorted samples were further processed for multiplex qPCR analysis. (b) PCA plot showing transcriptional signatures of quadruplicates of bulk cell samples (dots) at the indicated stages of WT Shigella infection (BY, VAC, and CYT) or of the uninfected control (CO). PC1 and PC2 and their percentages of captured variance are shown. The lines between samples indicate the two nearest neighbors. Representative data from one out of three independent experiments are shown (see Fig. S4a and b and Movies S1 to S3 in the supplemental material). (c) Heat map showing median ΔCT values of bulk cell samples transformed into a row Z-score of the indicated genes under the indicated conditions of Shigella WT infection (BY, VAC, and CYT) or of an uninfected control (CO). All genes with at least one significant gene expression change under one of the conditions are shown. The color key indicates the row Z-scores ranging from +2 (high expression; yellow) to −2 (low expression; blue). Data from two independent experiments at a P value cutoff of 0.05 are shown (see Table S2 in the supplemental material).