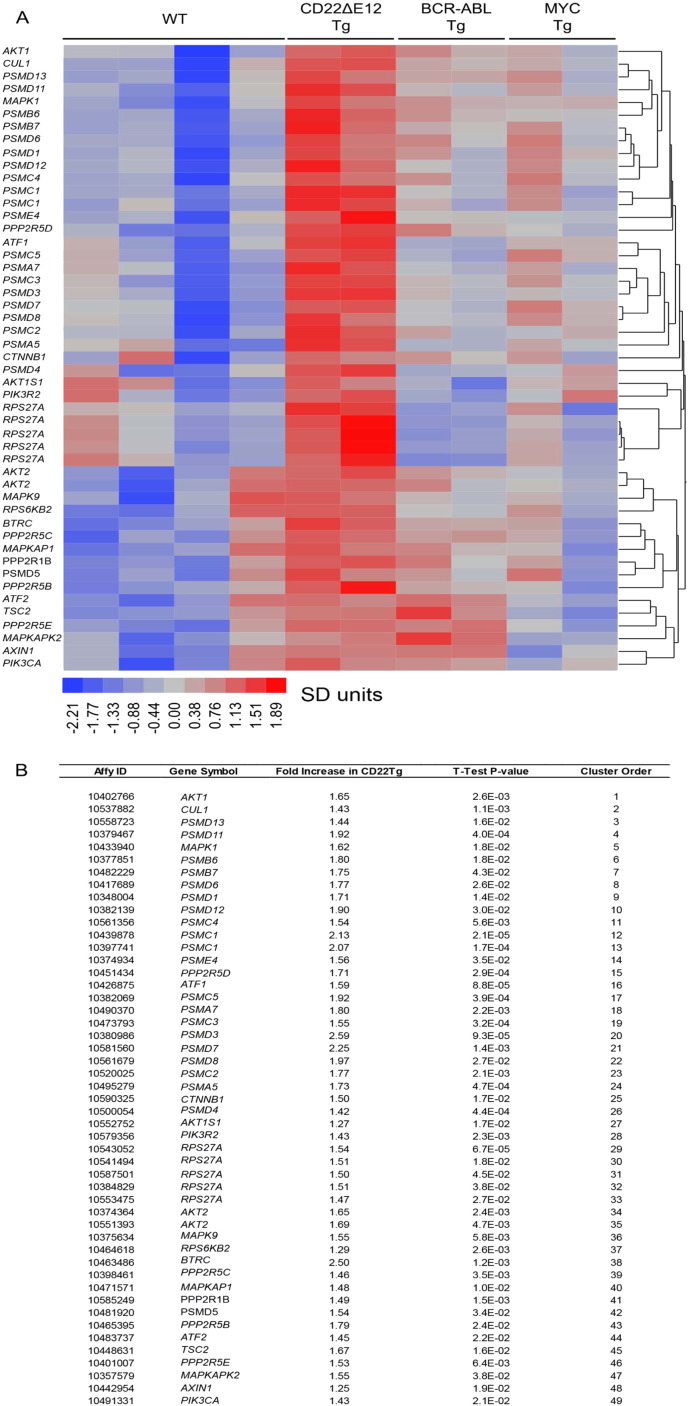
Fig. 2.
Signature transcriptome of CD22ΔE12 transgenic (Tg) BPL cells.
Gene expression values for splenocytes from WT healthy C57BL/6 mice (N = 4), leukemia cells from CD22ΔE12 Tg mice (N = 2), leukemia cells from BCR-ABL Tg mice (N = 2), and leukemia cells from MYC Tg mice (N = 2) were estimated from RMA normalization of signal values following hybridization to the Affymetrix Mouse Gene 1.0 ST Array (1102500 probes, 35512 genes). Expression values for each probeset were standardized across the 10 samples and differences in expression were visualized using a one-way hierarchical cluster figure to organize similar expression profiles of genes across the samples. Heat map depicts up- and downregulated transcripts ranging from red to blue respectively and clustered according to average distance metric (panel A). We determined a unique CD22ΔE12 signature transcriptome from significantly affected expression values comparing CD22ΔE12 Tg BPL cells with BPL cells from Eμ-MYC Tg or BCR-ABL Tg mice as well as WT splenocytes. T-tests documented that 49 probesets representing 43 signaling pathway genes in MAPK, PI3-K and WNT signaling networks were significantly upregulated in CD22ΔE12-Tg mice (P < 0.05).