Abstract
Engineering microbial diversity to enhance soil functions may improve the success of direct revegetation in sulphidic mine tailings. Therefore, it is essential to explore how remediation and initial plant establishment can alter microbial communities, and, which edaphic factors control these changes under field conditions. A long-term revegetation trial was established at a Pb-Zn-Cu tailings impoundment in northwest Queensland. The control and amended and/or revegetated treatments were sampled from the 3-year-old trial. In total, 24 samples were examined using pyrosequencing of 16S rRNA genes and various chemical properties. The results showed that the microbial diversity was positively controlled by soil soluble Si and negatively controlled by soluble S, total Fe and total As, implying that pyrite weathering posed a substantial stress on microbial development in the tailings. All treatments were dominated by typical extremophiles and lithotrophs, typically Truepera, Thiobacillus, Rubrobacter; significant increases in microbial diversity, biomass and frequency of organotrophic genera (typically Nocardioides and Altererythrobacter) were detected in the revegetated and amended treatment. We concluded that appropriate phytostabilization options have the potential to drive the microbial diversity and community structure in the tailings toward those of natural soils, however, inherent environmental stressors may limit such changes.
Sulphidic base metal tailings represent a typical anthropogenic extreme environment, as indicated by the much lower diversity of dwelling microbial communities relative to natural soils. During tailings phytostabilization, which aims to stabilize surface tailings through the establishment of a sustainable plant cover1, the microbial diversity and community structure are expected to be shifted by remediation measures and plants established. It is known that soil microbial diversity is critically associated to soil functioning, among which the most obvious one is to support plants2. Thus, the extent and direction of microbial structural shift in tailings is not only an indicator for soil development but also an essential factor for plant sustainability even in a short-term. Yet we still know very little about how phytostabilization practice changes tailings microbial community structure and what edaphic factors in tailings control the changes under field conditions.
Depending on the ore type, tailings can be quite diverse in their physicochemical properties, but can generally be described as sandy/silty, oligotrophic and toxic3. In base metal mines, the tailings are always sulphidic and abundant in residue metals and metalloids (mainly As)4,5. These tailings are normally dumped as impoundments devoid of vegetation. To reduce the adverse effects of legacy tailings, surface stabilisation through revegetation (i.e. phytostabilization) is legally required in many parts of the world. However, in contrast to phytoremediation of contaminated land6 or mine spoils7, phytostabilization of sulphidic base metal tailings is highly constrained by the inability of the tailings to support plants.
Microbial communities are essential players in soil functioning8 and biogeochemical cycling of tailings9. In contrast to acidic mine environments, the diversity and structure of microbial communities in neutral/alkaline tailings has not been extensively studied. Therefore, there had been little understanding of the dynamics of microbial communities during long-term field phytostabilization trials in extensively weathered sulphidic tailings in alkaline pH condition. Significant microbial biomass can be present in the oxidized layer of neutralised base metal tailings, but with lower microbial diversity (mainly bacteria) compared to natural soils9,10,11,12. A limited number of studies have also implied that the establishment of pioneer plants in Pb-Zn tailings increased microbial biomass and changed the community structure11,13. Nevertheless, how the microbial community structure in neutral base metal tailings changes with organic matter amendment and pioneer plant colonization, and the edaphic factors that drive these changes, are yet to be explored.
We hypothesize that in weathered neutral base metal tailings organic matter addition and colonization of pioneer plant species could substantially increase microbial diversity and change the microbial community structure. This study took advantage of a long-term field trial established at a Pb-Zn-Cu mine tailings impoundment in northwest Queensland, under subtropical and semi-arid climatic conditions. Tailings samples were examined for microbial diversity through pyrosequencing using universal primers for the 16S rRNA gene and a detailed analysis of environmental variables. Specifically, we aim to 1) examine the microbial diversity in the tailings on both revegetated and un-revegetated sites and 2) explore the edaphic factors controlling the evolution of the microbial community structure in the tailings. Our results suggest that organic matter remediation and plant establishment could induce the succession of microbial communities in the weathered and neutral tailings. However we believe that this succession is suppressed by geochemical factors (e.g. soluble S, total Fe and total As), which are the products of weathering of primary minerals in the tailings; these factors are retained in tailings in the semi-arid climatic conditions, where leaching is limited.
Results
The tailings were neutral (pH about 7), slightly/moderately saline (2–7 mS/cm; based on Australian classification), contained high levels of metals and metalloids (e.g. As, Zn), and low levels of nutrients (water soluble organic carbon (SOC) < 0.04%). SOC increased significantly in the woodchip amended and/or revegetated treatments, although other soil chemistries remained unchanged (Table 1).
Table 1. Selected properties of the 24 tailings samples used in this study.
| Treatment | Replicate and Code | Shannon index | OTUs number | EC (mS/cm) | pH | WC (%) | SOC | SON | HSOC | HSON | S-Sulfate | S-Nitrate (mg/kg) | S-P | S-Zn | T-As | T-Cu | T-P | T-Pb | T-Zn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tailings only | T_1 | 5.49 | 107 | 5.62 | 6.9 | 4.27 | 19.97 | 2.83 | 40.80 | 1.49 | 20870 | 0 | 3.51 | 13.63 | 217.2 | 1037 | 160.4 | 3620 | 1816 |
| T_2 | 4.28 | 123 | 6.68 | 6.9 | 2.66 | 37.47 | 3.86 | 44.54 | 1.52 | 29347 | 1.04 | 1.85 | 18.01 | 301.9 | 1230 | 230.4 | 5930 | 2910 | |
| T_3 | 5.02 | 166 | 4.70 | 6.7 | 4.50 | 8.96 | 2.09 | 31.25 | 1.15 | 16935 | 1.05 | 3.12 | 12.72 | 266.5 | 1464 | 233.9 | 5250 | 2532 | |
| T_4 | 3.94 | 92 | 2.26 | 6.8 | 12.73 | 11.11 | 2.22 | 38.16 | 1.23 | 13513 | 0 | 5.58 | 11.00 | 320.6 | 1312 | 319.3 | 6760 | 3224 | |
| T_5 | 5.07 | 111 | 3.22 | 7.4 | 12.79 | 11.37 | 2.43 | 40.20 | 1.46 | 11716 | 1.03 | 3.12 | 12.38 | 303.9 | 1382 | 330.8 | 6730 | 3409 | |
| T_6 | 4.68 | 105 | 2.89 | 6.8 | 12.63 | 23.24 | 2.49 | 40.09 | 1.23 | 8655 | 0.38 | 1.76 | 11.55 | 309.9 | 1253 | 259.3 | 6400 | 3134 | |
| Tailings revegetated | T+P_1 | 5.93 | 149 | 5.42 | 6.8 | 9.41 | 197.91 | 11.19 | 130.21 | 9.37 | 17521 | 6.11 | 1.67 | 9.42 | 286.3 | 1272 | 238.9 | 5300 | 3234 |
| T+P_2 | 5.67 | 200 | 2.59 | 6.9 | 12.93 | 43.58 | 11.49 | 72.71 | 5.83 | 14340 | 10.85 | 3.87 | 4.15 | 299.7 | 1334 | 330.1 | 6370 | 3579 | |
| T+P_3 | 5.14 | 95 | 2.08 | 6.9 | 9.85 | 19.18 | 6.92 | 78.14 | 5.83 | 19352 | 5.67 | 1.93 | 13.33 | 275.0 | 1275 | 308.9 | 5090 | 3164 | |
| T+P_4 | 4.71 | 78 | 4.59 | 6.8 | 1.07 | 49.27 | 38.99 | 74.01 | 20.27 | 15982 | 42.89 | 1.65 | 8.73 | 305.4 | 1308 | 281.7 | 5660 | 3275 | |
| T+P_5 | 5.56 | 181 | 5.89 | 6.9 | 1.15 | 95.97 | 56.79 | 83.65 | 21.85 | 27719 | 17.74 | 2.63 | 11.59 | 299.3 | 1299 | 254.8 | 5910 | 3300 | |
| T+P_6 | 4.91 | 84 | 5.51 | 6.9 | 1.42 | 78.60 | 62.10 | 86.78 | 17.39 | 14008 | 22.91 | 3.79 | 16.00 | 268.8 | 1249 | 249.6 | 5290 | 2938 | |
| Tailings with woodchips amendment | T+W_1 | 4.79 | 101 | 2.61 | 6.9 | 11.34 | 12.84 | 1.77 | 46.93 | 1.54 | 10357 | 0.40 | 7.06 | 8.79 | 296.4 | 1227 | 280.3 | 6490 | 3070 |
| T+W_2 | 4.94 | 107 | 3.12 | 6.7 | 10.97 | 53.46 | 4.24 | 74.81 | 4.30 | 9659 | 0 | 3.33 | 9.58 | 286.8 | 1255 | 265.7 | 6350 | 2991 | |
| T+W_3 | 5.29 | 139 | 2.86 | 6.9 | 10.26 | 22.14 | 2.71 | 61.24 | 3.05 | 14330 | 0 | 0.84 | 7.88 | 316.3 | 1260 | 265.5 | 6410 | 3192 | |
| T+W_4 | 3.89 | 98 | 3.26 | 6.9 | 9.87 | 29.76 | 3.16 | 53.01 | 1.88 | 10328 | 0 | 2.49 | 17.49 | 297.8 | 1114 | 265.2 | 6000 | 3287 | |
| T+W_5 | 4.94 | 132 | 3.04 | 6.8 | 10.65 | 32.89 | 4.25 | 53.59 | 3.12 | 18580 | 11.18 | 2.76 | 11.27 | 258.6 | 1166 | 260.0 | 5860 | 3261 | |
| T+W_6 | 4.82 | 124 | 3.39 | 6.8 | 11.04 | 20.67 | 3.18 | 74.50 | 3.52 | 12530 | 0.45 | 1.88 | 5.25 | 259.8 | 1334 | 334.7 | 6140 | 3485 | |
| Revegetated tailings with woodchips amendment | T+P+W_1 | 5.57 | 162 | 3.90 | 6.9 | 12.87 | 36.83 | 6.99 | 83.73 | 5.67 | 9582 | 0.88 | 4.86 | 5.48 | 213.2 | 1086 | 215.8 | 4200 | 2409 |
| T+P+W_2 | 6.08 | 150 | 5.49 | 6.7 | 11.42 | 82.69 | 14.64 | 96.33 | 5.96 | 14943 | 1.63 | 3.53 | 37.54 | 240.8 | 1052 | 322.6 | 5080 | 3914 | |
| T+P+W_3 | 4.54 | 138 | 3.86 | 6.9 | 13.00 | 45.21 | 9.04 | 71.69 | 5.87 | 10999 | 1.09 | 2.41 | 5.27 | 237.3 | 1029 | 236.0 | 4700 | 2620 | |
| T+P+W_4 | 5.89 | 244 | 3.86 | 6.8 | 10.31 | 29.50 | 22.67 | 77.03 | 11.78 | 5628 | 0 | 6.61 | 4.09 | 224.9 | 1144 | 259.8 | 4930 | 2589 | |
| T+P+W_5 | 5.83 | 212 | 3.81 | 6.7 | 8.76 | 47.91 | 8.19 | 69.10 | 5.34 | 12247 | 1.95 | 3.74 | 15.27 | 284.9 | 1240 | 315.6 | 6300 | 3189 | |
| T+P+W_6 | 6.03 | 133 | 2.99 | 7.0 | 11.34 | 47.95 | 11.69 | 83.91 | 6.12 | 6595 | 3.30 | 4.00 | 8.30 | 258.9 | 1046 | 250.9 | 5890 | 2772 |
Notes: WC, water content; EC, electrical conductivity; SOC, soluble organic carbon content; SON, soluble nitrogen content; HSOC, hot water soluble organic carbon content; HSON, hot water soluble nitrogen content; “S” and “T” as prefixes with elements/ions stand for soluble and total, respectively.
There was a large heterogeneity in the physicochemistry among the replicates in all treatments due to long-term uneven watering, root distribution and salt movement (Supplementary Table 1). For example, in the T+P treatment, EC ranged from 2.1 mS/cm to 5.9 mS/cm, water content ranged from 1.1% to 13.0% and SOC from 19.2 mg/kg to 198.0 mg/kg. Such heterogeneity means that the 24 samples are unique to each other, thus expanding the gradient ranges of physicochemical conditions in the tailing samples. For ecological statistics, the number of samples in this study is treated as 24.
In total, 40,988 good quality reads were obtained in all the samples with pyrosequencing. These sequences were classified into 557 OTUs0.97, including five archaeal. The number of OTUs ranged from 78 (T+P_4) to 244 (T+P+W_4). The T+P+W treatment had significantly (P = 0.019, one-way ANOVA; below the same) higher OTU numbers than the other treatments. The Shannon index ranged from 3.90 (T+W_4) to 6.08 (T+P+W_2) across all the samples (Table 1). Again, the Shannon index was significantly (P = 0.007) higher in T+P+W than the other treatments.
Rarefaction analysis of the microbial communities is shown in Supplementary Figure 1. At a 3% genetic distance, all the Shannon indices tended to be saturated against the sampled sequence number. This indicated that, despite some samples (9 of 24) producing a fairly low number of quality reads (<1000), the pyrosequencing survey covered almost the full extent of the taxonomic diversity in all the tailings samples.
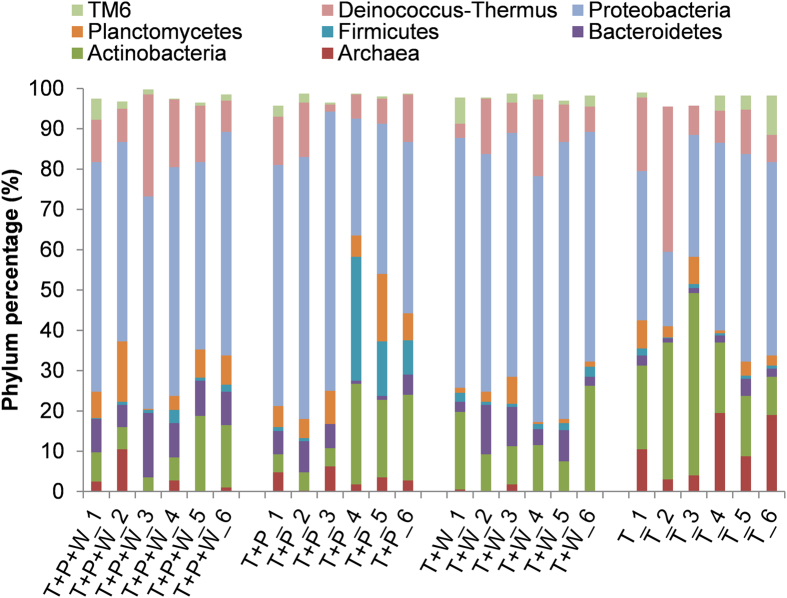
The 554 bacterial and archaeal OTUs0.97 were affiliated with 18 phyla. Across all 24 samples, Actinobac-teria, Bacteroidetes, Planctomycetes, Proteobacteria, Deinococcus-Thermus, and Crenarchaeota were found to be the dominant phyla. Among these, Actinobacteria, Proteobacteria and Deinococcus-Thermus accounted for >60% of all the sequences, and for >72% in all the averaged treatments (Fig. 1).
Figure 1. Phylum-level microbial composition of the tailings samples in this study.
The phyla with a low frequency (<3%) in all the samples, including Acidobacteria, Armatimonadetes, Chlamydiae, Cyanobacteria, TM7, SBR1093, OP11 and Chloroflexi, were not shown in this figure.
The OTUs were further classified into 36 classes (6 OTUs0.97 were not assigned by the Classifier) and 57 orders (71 OTUs0.97 were not assigned). The dominant classes across all samples were Alphaproteobacteria, Gammaproteobacteria, and Deinococci (>1% in all samples). When averaged treatment data was used, eight classes were found to be >1% in all treatments.
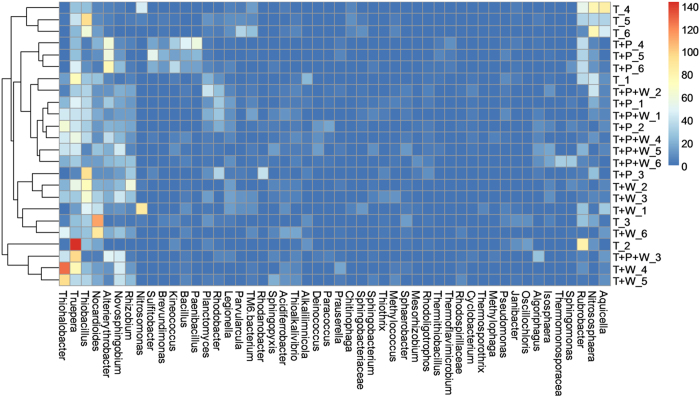
Abundant OTUs in each sample were selected and a heatmap was generated for comparison among the samples (Fig. 2 and Supplementary Figure 2 for phylogeny of major OTUs). Interestingly, on average, only one genus, Truepera, was found to be >5% and four other genera (Thiobacillus, Rubrobacter, Nocardioides and Altererythrobacter) were >1% in all four treatments. When the 24 individual samples were compared, only the five genera listed above were present in all samples.
Figure 2. A heatmap showing the comparison and cluster analysis of microbial composition in the 24 tailings samples in this study.
Indicator scores are based on the OTU abundance in the microbial assemblages. Only the OTUs with a frequency of >1% in at least 1 sample were shown in this figure.
Due to the large variation among the replicates, the differences in abundance of most phyla were not statistically significant among the four treatments, with the exception of T+P+W, which was significantly higher in Bacteroidetes (P = 0.001, as above); the tailings-only sample harboured considerably higher Archaea (P < 0.001) and significantly lower Proteobacteria (P < 0.001) than the other treatments. At the genus level, T+W contained significantly higher in Thiohalobacter (P = 0.006); the two revegetated treatments contained less Thiobacillus (P < 0.04) and Aquicella (P < 0.02) than the two non-revegetated; T+P contained significantly higher Altererythrobacter sp. than the others (P < 0.001).
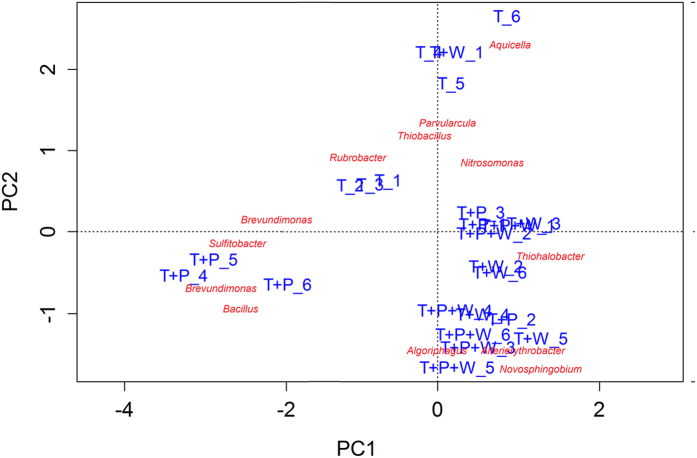
Microbial composition of the four treatments was differentiated by PCA at both the treatment (Supplementary Figure 3) and replicate levels (Fig. 3). The tailings-only treatment was clearly separated from the other treatments despite the substantial deviation among replicates. The tailings-only treatment was featured by a cluster of Rubrobacter, Aquicella, Nitrososphaera, Parvularcula, Thiobacillus and Nitrosomonas, while a cluster of Altererythrobacter, Bacillus, Paenibacillus, Algoriphagus and Thermomonosporaceae was dominant in the revegetated samples.
Figure 3. Biplots of the principal component analysis (PCA) at the replicate levels showing the correlation of genera with PCA axes.
The PCA axes differentiate the tailings samples according to their microbial composition.
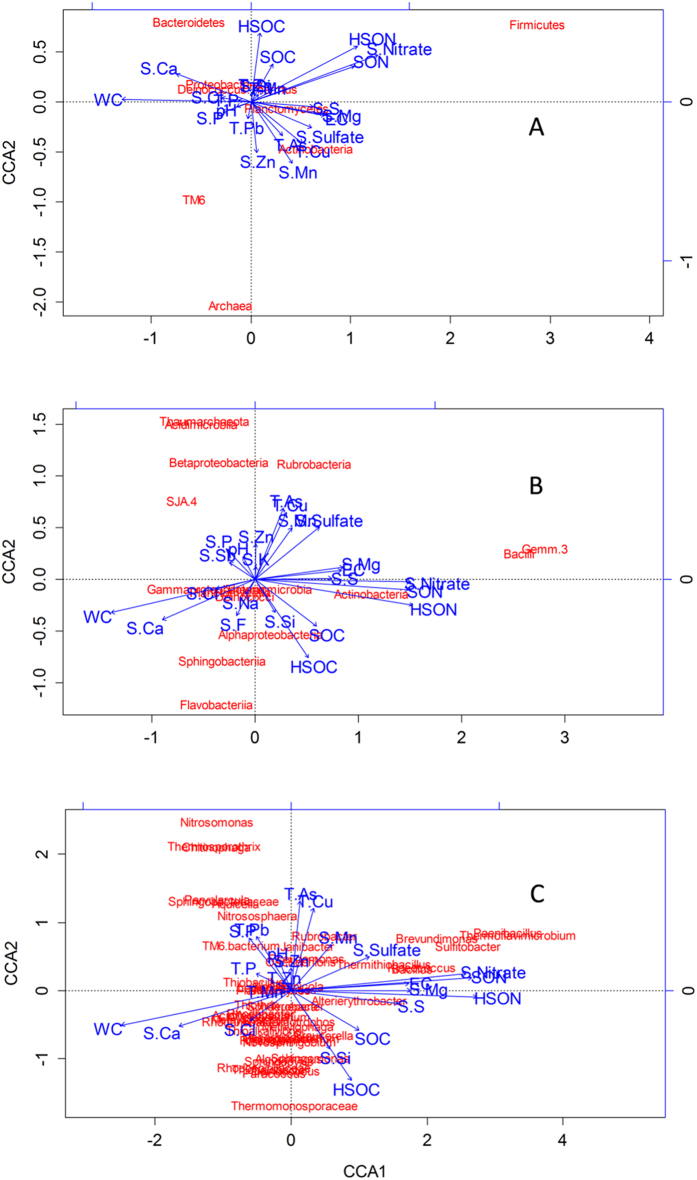
To explore which environmental factors best explain the microbial community structure, CCA was performed. A statistical analysis showed that significant correlations existed both between biological and environmental variables and among the biological or environmental variables (Supplementary Figure 4). Although HSOC was mainly controlled by SOC (r = 0.82) and SON (r = 0.43), the alpha diversity and Shannon index were both positively correlated with soluble Si (r = 0.63 and 0.60, respectively) and negatively correlated with soluble S (r = −0.28 and −0.31), total Fe (r = −0.31 and −0.51) and total As (r = −0.31 and −0.44).
CCA results at the phylum, class and genus levels indicated that the soil N status and water content were the best predictors for microbial community structure (Fig. 4). Hot water soluble N, soluble N and water content explained 49%, 38% and 45% of the variation observed in the microbial community structure, respectively. Together, these variables explained up to 53% of the variation (Rank correlation method: Spearman; Resemblance measure: D1 Euclidean distance).
Figure 4. Biplots of canonical correspondence analysis of taxa abundance Vs. environmental variables in the 24 tailings samples.
Three taxonomic levels, the phylum (A), the class (B) and the genus (C), were analysed. Phyla of maximum abundance <10%, classes of maximum abundance <3%, and genera of maximum abundance <1% in all the samples were excluded from the analysis. Abbreviations can be found in Table 1.
CCA further indicated that at the phylum level, Firmicutes was strongly associated with the soil N status (total soluble N, soluble NO3– and hot water soluble N), Archaea with soil soluble Zn, and Mn and Bacteroidetes with soluble organic C. At the class level, Rubrobacteria were strongly associated with soluble Zn and soluble sulphate. Bacilli were strongly associated with the soil N pool. At the genus level, some prominent associations were also found. Kineococcus, Paenibacillus and Bacillus were strongly associated with soil N status and Nitrososphaera was associated with total As.
Discussion
Structural shift of soil microbial communities under pressure from environmental factors is believed to have functional implications14. In the context of tailings revegetation, we expected that amendment and establishment of plants, even if only temporarily, could stimulate a change in the microbial community structure of the tailings towards a structure more similar to those of normal soils with sustainable biological capacity. Thus, exploring the factors controlling the microbial community structure in tailings in response to revegetation is important for assessing the efficacy of remediation measures and for gaining a better understanding of induced changes so superior remediation strategies can be developed.
As expected, the T+P+W treatment had significantly higher microbial diversity and biomass than the control, although the diversity and biomass in all treatments were still considerably lower than values reported for normal soils15,16. A substantial structural difference was observed between the control and treatments, with the emerging trend of increased organotroph-dominated microbes, compared to the lithotroph-dominated community in the tailings without amendment and plants. Yet surprisingly, soluble N, rather than SOC, was found to be the predominant factor driving the microbial structural evolution.
In the tailings used in this study, the concentrations of As, Cu, Pb and Zn were at least 101, 15, 362 and 23 fold higher, respectively, than the average crustal abundance17. Statistically, these elements are common stressors in Pb-Zn mine tailings worldwide (Supplementary Figure 5). High salinity is also of concern in the tailings, which would persist in the root zone profile due to the high geochemical reaction potential of tailings minerals and poor leaching in the semi-arid environment3. Though the tailings can be classified as moderately saline based on the data in this study, the pore water EC has reached values of up to 51.3 mS/cm over the past three years3. Moreover, nutrient deficiency is a common constraint for microorganisms living in tailings. An initial comparison (climatic factors were not considered) showed that the average SOC of the tailings samples was far less than the values reported for natural and arable soils18,19,20 and close to those of highly contaminated soils21.
Reflecting the stressors in this environment, diverse extremophiles were abundant in the tailings. At the treatment level, two potential extremophile genera, Truepera and Rubrobacter, were dominant and were shared by all treatments. The genera Truepera, within the Deinococcales group, and Rubrobacter affiliated to phylum Actinobacteria, both contain several well-characterized thermophilic radiation-tolerant species22,23,24,25. Due to the extreme DNA damage resistance inherent in these genera, they are possibly well-adapted to the oxidative conditions of the tailings caused by seasonal drought26, high levels of toxic elements27 and salinity28. Some other extremophiles, including the radiation-tolerant genera Kineococcus (>1% in eight samples)29 and Deinococcus (>1% in 11 samples)30 and a halophile genus Thiohalobacter (>1% in 15 samples)31, were also abundant in the tailings. These known radiation-tolerant, halophile, thermophile and/or metal-tolerant genera accounted for 27% (T+W_1) to 71% (T_2) (averagely 42%) of OTUs in all microbial assemblages.
A spectrum of potential S- and/or Fe-oxidizing genera was abundant in all samples. These included Thiobacillus32,33, Thiohalobacter31, Sulfitobacter34,35,36, Acidiferrobacter37, Thioalkalivibrio38,39,40, Alkalilimnicola41, and Thiothrix 42. These genera accounted for 6% to 42% (average 20%) of OTUs across the 24 microbial assemblages. The existence of abundant S- and/or Fe-oxidizing phylotypes using pyrites as a major energy source has been reported in many sulphidic tailings worldwide9,43,44,45. This again highlights the role of pyrites in shaping microbial communities associated with sulphidic tailings.
In terrestrial primary succession, higher plants only appear when soil properties reach a critical point46,47, with microbial diversity being one of the key soil properties48,49. Although abiotic factors pose significant constraints for plant establishment in mine tailings50, the development of microbial diversity is also crucial to facilitate and sustain plant growth and therefore must be part of the engineering efforts. Despite the overall low microbial biomass and alpha-diversity in all samples, this study found a substantial structural progression of microbial communities in response to phytostabilization (Supplementary Figure 3). Key changes from the tailings only to the revegetated treatments included the decrease of Nitrososphaera, Rubrobacter, TM6 bacteria and Aquicella and the increase of Novosphingobium and Altererythrobacter. The latter two genera are reported degraders of complex organic matter, for example, cellulose or aromatic compounds51,52,53. Further, the increase of heterotrophic species (mostly Bacteroidetes and Proteobacteria, e.g. Rhizobium and Rhodobacter) in the revegetated treatments was a major contributor to the increase in the Shannon index (data not shown). In natural soils, Bacteroidetes and Proteobacteria are among the most abundant phyla54 and are statistically associated with soil C turnover55. Many studies have shown that terrestrial microbial succession from non-vegetated to vegetated environments follows predictable patterns due to the convergence effects of plants56. Therefore, although mine tailings have a distinct initial microbial composition compared to other soil-formation materials like volcanic ash57 and glacier forefronts58,59, we speculate that phytostabilization may induce a directional change in the microbial community structure from autotroph- to heterotroph-dominant in sulphidic tailings, resembling natural primary succession60,61.
It is agreed in the literature that the driving factors for the evolution of a microbial community structure largely depend on the environmental gradient to which the communities are subjected or imposed under experimentally62. Organic C63, N availability64, pH65, salinity66,67, moisture68 and many other abiotic factors influence the microbial community structure in soil. Despite this, the edaphic gradients were not found to equally impact the community structure in the tailings of this study, with soluble N and water content being the best controllers. The predominant role of N in controlling the structure of the tailings microbial community is similar to findings in terrestrial primary succession, which indicated that N is a primary limiting factor at early stages of soil development47. In the tailings where SOC and SON were low and metal stress was high, Nitrososphaera were found to be dominant (Fig. 4A). Species within Nitrososphaera69,70 are typical ammonia-oxidizers well-adapted to a heavy metal-rich environment. They may have found a niche in the tailings and play a major role in ammonium-N oxidation. As significant nitrification needs sufficient ammonium-N71, the existence of abundant Nitrososphaera may also indicate active N fixation in these tailings samples, which was possibly performed by the other predominant genera Rubrobacter (CP000386 in Genbank) and Truepera72 that may harbour nitrogenases. Firmicutes were shown to respond positively to soluble N (Fig. 4A). Many genera within Firmicutes, such as Paenibacillus and Bacillus, are well-known degraders; some have been used as plant growth promoting rhizobacteria73. Thus, we found N availability to be a primary factor shaping the microbial communities associated with the tailings. For phytostabilization of the tailings, the addition of soluble N may shift the microbial community structure toward an increase in heterotrophs and could ‘prime’ nutrient cycling in the tailings.
It should be noted that all the associations discussed above are based on the premise that the microbial communities were shaped mainly by deterministic processes, specifically that habitat filtering effects are the dominant drivers in determining microbial composition in the tailings. Some random processes that are not accounted for here could also be important74. In this study, these may include competition among species in similar niches, as reflected by some of the negative associations detected among similar genera, and seed availability, which can be different among treatments by the introduction of plants or amendments. Meanwhile, once-off sampling may be not adequate to draw a reliable relationship between environmental factors and microbial community structure, since both of many factors, like water content and labile N, and microbial communities are dynamic under field conditions. Yet, as an extreme environment, sulphidic tailings may have strong selective effects on colonizing species, even if organic amendments and plant roots can create microhabitats, as shown in this study. To gain further understanding of the dynamic evolution of the microbial communities under the site conditions, it is useful to conduct repeated sampling from the same location in multiple seasons in the near future.
As expected, fairly low microbial biomass (i.e. hot water soluble carbon) and alpha-diversity (i.e. the number of OTUs0.97) were found in all the samples. This concurs with the ecological amplitude theory, which predicts that extreme habitats usually have low biomass and biodiversity relative to moderate habitats14,75,76. The average HSOC of the tailings in this study was less than 1/5 of the reported values for grassland and forest soils20,77. The number of microbial OTUs in the tailings obtained through in-depth tag-sequencing was close to those found in other tailings using 16S rDNA clone libraries. For example, around 160 OTUs0.99 were found at a Pb-Zn mine site in Graham County, Arizona, USA43; 23–50 OTUs0.97 were found in the Pb-Zn tailings in Yunnan, China44,45; and 86 OTUs0.97 were found in the Pb-Zn tailings in Guangdong, China78. For comparison, forest and grassland soils have considerably higher microbial alpha-diversity (around 3000 OTU0.97)79.
In contrast to natural pedogenesis, tailings contain a high abundance of stressors that may not be removed sufficiently and rapidly enough via natural succession61. In our semi-arid site, the removal of these stressors (soluble salts from mineral weathering) is exacerbated due to the lack of frequent water infiltration and very limited leaching processes. The negative role of pyrites (FeS2) in mine tailings in controlling microbial diversity has been indicated by the negative correlations between the alpha diversity plus Shannon index and total Fe, soluble S plus total As. While previous studies have confirmed that the tailings in this study are abundant in pyrites80, the soluble S can be an indicator of active pyrite weathering81,82. Active weathering of pyrite will release not only saline ions by dissolving carbonates, but also toxic elements, especially As, as various sulphides normally associate with pyrite as sulphide paragenesis83. Indeed, Fe was strongly correlated with total As (r = 0.73), total Cu (r = 0.66), total Mn (r = 0.78), total Zn (r = 0.54) and total Pb (r = 0.61) in this study. Meanwhile, as estimated in many studies81,84, pyrite oxidation can continue for hundreds of years in most pyrite-abundant tailings. From an ecological engineering perspective, this means that phytostabilization of tailings is not practical for creating a safe site for either belowground or aboveground communities due to the long-term release of saline or metal/metalloid elements. Therefore, the roles of pyrite-eaters, metal-tolerant and halophile genera in soil functioning merit further attention.
Conclusions
Phytostabilization of sulphidic tailings remains a challenging enterprise. As indicated by the results presented in this study, the direct revegetation of tailings with amendment of woodchips induced significant increases in both microbial diversity and biomass and directed the microbial community structure towards one more similar to those of natural soils. Those changes, however, did not modify the tailings sufficiently to meet the requirements of a functioning soil. The low microbial diversity and biomass of the communities and the dominance of extremophiles in all the treatments means additional inputs and on-going improvement would be required, in addition to the initial amendment and direct revegetation with pioneer plant species, although the addition of N fertilizers and a water regime could be used to ‘manipulate’ microbial communities. Considering that rapid removal of most stressors in tailings via leaching is not a practical option under semi-arid climatic conditions, searching for more efficient amendments to ameliorate the tailings is a priority for the phytostabilization of tailings85,86.
Materials and Methods
Sampling
The field trial site was located at a Pb-Zn-Cu mine tailings impoundment in northwest Queensland, with a subtropical and semi-arid climate with intensive rainfall in summer. The mine had been in operation as a Pb-Zn-Ag and/or Cu mine for over 80 years and was decommissioned about 30 years ago. Accumulated mine tailings in this area were estimated to be over 100 million tonnes80.
The field trial was established in 2010 to investigate options for direct revegetation of the tailings, which had experienced 40 years of natural weathering. Briefly, four treatments were designed as follows: tailings-only as control (T), tailings directly revegetated with native plants (T+P) (see below), tailings with woodchip amendment (T+W) and tailings with woodchip amendment and directly revegetated with native plants (T+P+W). The woodchips were woody mulch from local trees. A total of 2 m2 plots were established for each treatment. In the treatments amended with woodchips, 20% (v:v) of the total volume of woodchips was amended into the tailings. Plants established in the tailing treatments were mixed native species (with 5–6 plants each species), including Acacia chisolmii, A. ligulata, Atriplex nummularia, and Ptilotus exaltatus. These were transplanted as tubestock seedlings. More information about treatment and plant status can be found in Supplementary Table 1. The field trial was initially established to compare main effects of these amendment approaches, but extreme field conditions resulted in high variability even within treatments. As a result, the data from subsamples were pooled for correlation analysis rather than simple treatment comparison, to explore major edaphic factors associated with microbial community profile.
Tailings were sampled in June 2013. Six separate subsamples were taken at six randomly distributed spots across the 2 m2 surface area of each treatment. This sampling method resulted in six quasi-replicates for each treatment. And considering the environmental heterogeneity in the field (as characterized by the physiochemistry of the samples below), the 24 subsamples have been treated ecologically as separate sample units for statistical analyses, in a similar manner as a survey. Samples were placed into double sealed plastic bags in a pre-cooled icebox (<10 °C) and shipped to the laboratory within 24 h. During DNA extraction, samples were stored in a cold room (4 °C) and were extracted within eight days. Aliquots of each replicate subsample were oven-dried for physiochemical analyses.
Physicochemistry
For total elemental analysis, ball-milled tailings samples were digested with a 5:3:2 solution of nitric, hydrochloric and hydrofluoric acids using a microwave system87. Total elements after digestion were then analysed using ICP-AES. Water soluble elements were extracted by shaking 1 g sieved tailings (2 mm) in 50 ml DI water for 1 h at room temperature4. The elements in the water extracts were then determined using ICP-AES after filtering through a 0.45 μm filter. pH and electrical conductivity (EC) were determined electrically with a soil/water ratio of 1:5. Soluble anions in the water extracts were determined using an ion chromatograph DX-120 (Dionex Corporation) equipped with Column AS-22. Water and hot water soluble C and N were analysed following the methods described by Ghani et al.77. Briefly, 3 g dry tailings were shaken with 30 ml distilled water for 30 min at room temperature. After centrifugation at 3500 rpm for 20 min, the supernatant was filtered through 0.45 μm filter for water soluble C and N analyses. A further 30 ml of water was added to the sediments and the tubes were shaken for 16 h in a water bath at 80 °C. Following further centrifugation, the supernatant was collected as described above for C and N analyses. C and N in the solutions were determined using a Shimadzu Total Organic Carbon Analyser with Total Nitrogen detector. Hot water soluble organic C was used as a proxy of microbial biomass77.
DNA extraction
DNA extraction from the tailings samples was performed following the protocol established in our previous study88 with minor modifications. Briefly, 5 g of tailings together with 25 ml 0.2% sodium pyro-phosphate solution (pH 7.5) were added into a 50 ml Falcon® tube and sonicated (2 × 30 s at moderate power output). The suspensions were immediately layered into a sucrose solution (25 ml of 1.3 g ml−1) in another 50 ml Falcon® tube, then centrifuged at 5,500 × g for 2 min. The clear upper sucrose fraction containing the microbial cells was poured into a new 50 ml Falcon® tube and centrifuged at 20,000 × g for 10 min at 4 °C. The top 1/3 supernatant was discarded and then the same volume of 0.8% NaCl solution was added to the tube and concentrated by centrifugation (20,000 × g for 10 min) at 4 °C. After discarding the supernatant, the cells plus fine particulates at the bottom of the tube were carefully rinsed with bead solution and transferred into the Bead Tube of PowerLyzerTM PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc.) for DNA extraction.
The concentration and purity of the extracted DNA was determined using a Nanodrop spectrometer (Thermo Scientific, US). Quality DNA was submitted to the Australian Centre for Ecogenomics (ACE) within The University of Queensland for Pyrosequencing, using universal fusion primers 926F and 1392wR as described elsewhere89.
Pyrosequencing data analysis
Pyrosequencing-derived data was analysed through the ACE pyrotag pipeline90. Briefly, after the data were ordered using QIIME91, the sequence reads were trimmed to 250 bp lengths and de-noised using ACACIA92. Sequences with 97% similarity were assigned to operational taxonomic units (OTUs0.97) by CD-HIT-OTU93 and aligned by Pynast91. A non-normalized OTU table and rarefaction curves were then generated by QIIME. A centroid normalized OTU table was generated using Nomaliser94. The normalized OTU table was used for further analyses.
Statistics
A heatmap showing the abundance of major genera was generated using the package ‘pheatmap’95 in R version 2.15.1. These OTU sequences and reference sequences from Genbank were aligned using SINA96. The aligned sequences were then used to generate a neighbour-joining phylogenetic tree using MEGA 597.
Linear correlation analysis (LCA), principal component analysis (PCA) and canonical correspondence analysis (CCA) were performed between the environmental variables and biological composition using R with the ‘vegan’ package98.
Additional Information
How to cite this article: Li, X. et al. From lithotroph- to organotroph-dominant: directional shift of microbial community in sulphidic tailings during phytostabilization. Sci. Rep. 5, 12978; doi: 10.1038/srep12978 (2015).
Supplementary Material
Acknowledgments
We thank Dr Fiona May of the Australian Centre for Ecogenomics, The University of Queensland for her work on pyrosequencing of the samples and Mr Yang Lu of the Advanced Water Management Centre, The University of Queensland for his help in processing the data. This project is financially supported by UQ Postdoctoral Fund and Mount Isa Mines, Glencore Ltd (formerly Xstrata Copper Ltd.).
Footnotes
Author Contributions L.H., P.B. and X.L. designed the experiment. X.L. and L.H. performed the experiment and X.L. analysed the data. X.L. wrote the first draft of the manuscript. J.V.N. and J.Z. helped in the interpretation of results. All authors contributed to the revisions.
References
- Li X. & Huang L. Toward a new paradigm for tailings phytostabilization – nature of the substrates, amendment options and anthropogenic pedogenesis. Cri Rev Env Sci Tec 45, 813–839 (2014). [Google Scholar]
- Foth H. D. Fundamentals of soil science (John Wiley & Sons, Inc., 1990). [Google Scholar]
- Huang L. B., Baumgartl T. & Mulligan D. Is rhizosphere remediation sufficient for sustainable revegetation of mine tailings? Ann Bot-London 110, 223–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dold B. Speciation of the most soluble phases in a sequential extraction procedure adapted for geochemical studies of copper sulfide mine waste. J Geochem Explor 80, 55–68 (2003). [Google Scholar]
- Yang Z. Y., Yuan J. G., Xin G. R., Chang H. T. & Wong M. H. Germination, growth, and nodulation of Sesbania rostrata grown in Pb/Zn mine tailings. Environ Manage 21, 617–622 (1997). [DOI] [PubMed] [Google Scholar]
- Cunningham S. D., Berti W. R. & Huang J. W. W. Phytoremediation of contaminated soils. Trends Biotechnol 13, 393–397 (1995). [Google Scholar]
- Li X. F., Park J. H., Edraki M. & Baumgartl T. Understanding the salinity issue of coal mine spoils in the context of salt cycle. Environ Geochem Hlth 36, 453–465 (2014). [DOI] [PubMed] [Google Scholar]
- Doran J. W. & Parkin T. B. in Defining soil quality for a sustainable environment (eds Doran J. W. et al.) 3–21 (Soil Sci. Soc. Am., Inc. and Am. Soc. Agron., Inc., Madison, WI. 1994).
- Diaby N. et al. Microbial communities in a porphyry copper tailings impoundment and their impact on the geochemical dynamics of the mine waste. Environ Microbiol 9, 298–307 (2007). [DOI] [PubMed] [Google Scholar]
- De la Iglesia R., Castro D., Ginocchio R., van der Lelie D. & Gonzalez B. Factors influencing the composition of bacterial communities found at abandoned copper-tailings dumps. J Appl Microbiol 100, 537–544 (2006). [DOI] [PubMed] [Google Scholar]
- Mendez M. O., Glenn E. R. & Maier R. M. Phytostabilization potential of quailbush for mine tailings: Growth, metal accumulation, and microbial community changes. J Environ Qual 36, 245–253 (2007). [DOI] [PubMed] [Google Scholar]
- Rosario K. et al. Bacterial community changes during plant establishment at the San Pedro River mine tailings site. J Environ Qual 36, 1249–1259 (2007). [DOI] [PubMed] [Google Scholar]
- Iverson S. L. & Maier R. M. Effects of compost on colonization of roots of plants grown in metalliferous mine tailings, as examined by fluorescence in situ hybridization. Appl Environ Microb 75, 842–847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A. Microbial community structure and its functional implications. Nature 459, 193–199 (2009). [DOI] [PubMed] [Google Scholar]
- Nedved V., Balik J., Cerny J., Kulhanek M. & Balikova M. The changes of soil nitrogen and carbon contents in a long-term field experiment under different systems of nitrogen fertilization. Plant Soil Environ 54, 463–470 (2008). [Google Scholar]
- Will C. et al. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl Environ Microb 76, 6751–6759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks D. L. Environmental soil chemistry (Academic Press, California, USA 2003). [Google Scholar]
- Lu X. Y., Fan J. H., Yan Y. & Wang X. D. Soil water soluble organic carbon under three alpine grassland types in Northern Tibet, China. Afr J Agr Res 6, 2066–2071 (2011). [Google Scholar]
- Ma X. Z. et al. Soil glycosidase activities and water soluble organic carbon under different land use types. J Soil Sci Plant Nut 10, 93–101 (2010). [Google Scholar]
- Wang Q. K. & Wang S. L. Response of labile soil organic matter to changes in forest vegetation in subtropical regions. Appl Soil Ecol 47, 210–216 (2011). [Google Scholar]
- Scaglia B. & Adani F. Biodegradability of soil water soluble organic carbon extracted from seven different soils. J Environ Sci-China 21, 641–646 (2009). [DOI] [PubMed] [Google Scholar]
- Albuquerque L. et al. Truepera radiovictrix gen. nov., sp nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol Lett 247, 161–169 (2005). [DOI] [PubMed] [Google Scholar]
- Carreto L. et al. Rubrobacter xylanophilus sp nov: A new thermophilic species isolated from a thermally polluted effluent. Int J Syst Bacteriol 46, 460–465 (1996). [Google Scholar]
- Chen M. Y. et al. Rubrobacter taiwanensis sp nov., a novel thermophilic, radiation-resistant species isolated from hot springs. Int J Syst Evol Micr 54, 1849–1855 (2004). [DOI] [PubMed] [Google Scholar]
- Suzuki K., Collins M. D., Iijima E. & Komagata K. Chemotaxonomic characterization of a radiotolerant bacterium, Arthrobacter radiotolerans - description of Rubrobacter radiotolerans gen nov, comb nov. FEMS Microbiol Lett 52, 33–39 (1988). [Google Scholar]
- Moran J. F. et al. Drought Induces oxidative stress in pea-plants. Planta 194, 346–352 (1994). [Google Scholar]
- Valko M., Morris H. & Cronin M. T. D. Metals, toxicity and oxidative stress. Curr Med Chem 12, 1161–1208 (2005). [DOI] [PubMed] [Google Scholar]
- Gueta Dahan Y., Yaniv Z., Zilinskas B. A. & Ben Hayyim G. Salt and oxidative stress: Similar and specific responses and their relation to salt tolerance in Citrus. Planta 203, 460–469 (1997). [DOI] [PubMed] [Google Scholar]
- Bagwell C. E. et al. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. Plos One 3, e3878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White O. et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571–1577 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Kovaleva O. L., Tourova T. P. & Muyzer G. Thiohalobacter thiocyanaticus gen. nov., sp nov., a moderately halophilic, sulfur-oxidizing gammaproteobacterium from hypersaline lakes, that utilizes thiocyanate. Int J Syst Evol Micr 60, 444–450 (2010). [DOI] [PubMed] [Google Scholar]
- Beller H. R. et al. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitfificans. J Bacteriol 188, 1473–1488 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolle J. L. C., Simmons S., Bathe S. & Norris P. R. Ferrous iron oxidation and rusticyanin in halotolerant, acidophilic ‘Thiobacillus prosperus’. Microbiol-Sgm 155, 1302–1309 (2009). [DOI] [PubMed] [Google Scholar]
- Long C., Lu X. L., Gao Y., Jiao B. H. & Liu X. Y. Description of a Sulfitobacter strain and its extracellular cyclodipeptides. Evid-Based Compl Alt 2011, 1–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. R. et al. Sulfitobacter litoralis sp nov, a marine bacterium isolated from the East Sea, Korea. Int J Syst Evol Micr 57, 692–695 (2007). [DOI] [PubMed] [Google Scholar]
- Pukall R. et al. Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the alpha-Proteobacteria. Int J Syst Bacteriol 49, 513–519 (1999). [DOI] [PubMed] [Google Scholar]
- Hallberg K. B., Hedrich S. & Johnson D. B. Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 15, 271–279 (2011). [DOI] [PubMed] [Google Scholar]
- Muyzer G. et al. Complete genome sequence of “Thioalkalivibrio sulfidophilus” HL-EbGr7. Stand Genomic Sci 4, 23–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G. et al. Complete genome sequence of Thioalkalivibrio sp. K90mix. Stand Genomic Sci 5, 341–355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Lysenko A. M., Mityushina L. L. & Kuenen J. G. Thioalkalivibrio thiocyanoxidans sp nov. and Thioalkalivibrio paradoxus sp nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria capable of growth on thiocyanate, from soda lakes. Int J Syst Evol Micr 52, 657–664 (2002). [DOI] [PubMed] [Google Scholar]
- Hoeft S. E. et al. Alkalilimnicola ehrlichii sp nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Micr 57, 504–512 (2007). [DOI] [PubMed] [Google Scholar]
- Chernousova E. et al. Thiothrix caldifontis sp nov and Thiothrix lacustris sp nov., gammaproteobacteria isolated from sulfide springs. Int J Syst Evol Micr 59, 3128–3135 (2009). [DOI] [PubMed] [Google Scholar]
- Mendez M. O., Neilson J. W. & Maier R. M. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microb 74, 3899–3907 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. B., Shi W., Yang M. X., Sha T. & Zhao Z. W. Bacterial diversity at different depths in lead-zinc mine tailings as revealed by 16S rRNA gene libraries. J Microbiol 45, 479–484 (2007). [PubMed] [Google Scholar]
- Zhang H. B. et al. Bacterial diversity in mine tailings compared by cultivation and cultivation-independent methods and their resistance to lead and cadmium. Microb Ecol 54, 705–712 (2007). [DOI] [PubMed] [Google Scholar]
- Alvarez-Molina L. L., Martinez M. L., Perez-Maqueo O., Gallego-Fernandez J. B. & Flores P. Richness, diversity, and rate of primary succession over 20 year in tropical coastal dunes. Plant Ecol 213, 1597–1608 (2012). [Google Scholar]
- Chapin F. S., Walker L. R., Fastie C. L. & Sharman L. C. Mechanisms of primary succession following deglaciation at Glacier Bay, Alaska. Ecol Monogr 64, 149–175 (1994). [Google Scholar]
- Aikio S., Vare H. & Strommer R. Soil microbial activity and biomass in the primary succession of a dry heath forest. Soil Biol Biochem 32, 1091–1100 (2000). [Google Scholar]
- Tscherko D., Hammesfahr U., Zeltner G., Kandeler E. & Bocker R. Plant succession and rhizosphere microbial communities in a recently deglaciated alpine terrain. Basic Appl Ecol 6, 367–383 (2005). [Google Scholar]
- Mendez M. O. & Maier R. M. Phytostabilization of mine tailings in arid and semiarid environments - An emerging remediation technology. Environ Health Persp 116, 278–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. P., Wang B. J., Liu Y. H. & Liu S. J. Novosphingobium taihuense sp nov., a novel aromatic-compound-degrading bacterium isolated from Taihu Lake, China. Int J Syst Evol Micr 55, 1229–1232 (2005). [DOI] [PubMed] [Google Scholar]
- Sohn J. H., Kwon K. K., Kang J. H., Jung H. B. & Kim S. J. Novosphingobium pentaromativorans sp nov., a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int J Syst Evol Micr 54, 1483–1487 (2004). [DOI] [PubMed] [Google Scholar]
- Xue X. Q. et al. Altererythrobacter xinjiangensis sp nov., isolated from desert sand, and emended description of the genus Altererythrobacter. Int J Syst Evol Micr 62, 28–32 (2012). [DOI] [PubMed] [Google Scholar]
- Roesch L. F. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1, 283–290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Bradford M. A. & Jackson R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007). [DOI] [PubMed] [Google Scholar]
- Fierer N., Nemergut D., Knight R. & Craine J. M. Changes through time: Integrating microorganisms into the study of succession. Res Microbiol 161, 635–642 (2010). [DOI] [PubMed] [Google Scholar]
- Fujimura R. et al. Analysis of early bacterial communities on volcanic deposits on the island of Miyake (Miyake-Jima), Japan: a 6-year study at a fixed site. Microbes Environ 27, 19–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. P. & Jumpponen A. Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol Ecol 23, 481–497 (2013). [DOI] [PubMed] [Google Scholar]
- Ohtonen R., Fritze H., Pennanen T., Jumpponen A. & Trappe J. Ecosystem properties and microbial community changes in primary succession on a glacier forefront. Oecologia 119, 239–246 (1999). [DOI] [PubMed] [Google Scholar]
- Mcintosh R. P. & Odum E. P. Ecological succession. Science 166, 403–404 (1969). [DOI] [PubMed] [Google Scholar]
- Odum E. P. The strategy of ecosystem development. Science 164, 262–70 (1969). [DOI] [PubMed] [Google Scholar]
- Fierer N. et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. PNAS 109, 21390–21395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P., Kandeler E. & Marschner B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol Biochem 35, 453–461 (2003). [Google Scholar]
- Cusack D. F., Silver W. L., Torn M. S., Burton S. D. & Firestone M. K. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 92, 621–632 (2011). [DOI] [PubMed] [Google Scholar]
- Lauber C. L., Hamady M., Knight R. & Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronov E. E. et al. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eurasian Soil Sci+ 45, 147–156 (2012). [Google Scholar]
- Wichern J., Wichern F. & Joergensen R. G. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 137, 100–108 (2006). [Google Scholar]
- Brockett B. F. T., Prescott C. E. & Grayston S. J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem 44, 9–20 (2012). [Google Scholar]
- Spang A. et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14, 3122–3145 (2012). [DOI] [PubMed] [Google Scholar]
- Tourna M. et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. PNAS 108, 8420–8425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahrawat K. L. Factors affecting nitrification in soils. Commun Soil Sci Plan 39, 1436–1446 (2008). [Google Scholar]
- Ivanova N. et al. Complete genome sequence of Truepera radiovictrix type strain (RQ-24(T)). Stand Genomic Sci 4, 91–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepecky R. A. & Hemphill H. E. The genus Bacillus-nonmedical. Prokaryotes 4, 530–562 (2006). [Google Scholar]
- Cornwell W. K., Schwilk D. W. & Ackerly D. D. A trait-based test for habitat filtering: Convex hull volume. Ecology 87, 1465–1471 (2006). [DOI] [PubMed] [Google Scholar]
- Terbraak C. J. F. & Gremmen N. J. M. Ecological amplitudes of plant-species and the internal consistency of ellenberg indicator values for moisture. Vegetatio 69, 79–87 (1987). [Google Scholar]
- Wall D. H. & Virginia R. A. Controls on soil biodiversity: Insights from extreme environments. Appl Soil Ecol 13, 137–150 (1999). [Google Scholar]
- Ghani A., Dexter M. & Perrott K. W. Hot-water extractable carbon in soils: A sensitive measurement for determining impacts of fertilisation, grazing and cultivation. Soil Biol Biochem 35, 1231–1243 (2003). [Google Scholar]
- Huang L. N. et al. Spatial and temporal analysis of the microbial community in the tailings of a Pb-Zn mine generating acidic drainage. Appl Environ Microb 77, 5540–5544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacke H. et al. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLOS One 6, e17000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth B., Edraki M. & Baumgartl T. Understanding the Long-Term Seepage Chemistry of Base Metal Mine Tailings in a Semi-Arid Tropical Climate, Mount Isa, Australia. Paper presented at 7th Australian workshop on acid and metalliferous drainage, Darwin. Indooroopilly, QLD, Australia: JKTech. (2011, June 21-24).
- Dold B. & Fontbote L. Element cycling and secondary mineralogy in porphyry copper tailings as a function of climate, primary mineralogy, and mineral processing. J Geochem Explor 74, 3–55 (2001). [Google Scholar]
- Rimstidt J. D. & Vaughan D. J. Pyrite oxidation: A state-of-the-art assessment of the reaction mechanism. Geochim Cosmochim Ac 67, 873–880 (2003). [Google Scholar]
- Pal D. C., Barton M. D. & Sarangi A. K. Deciphering a multistage history affecting U-Cu(-Fe) mineralization in the Singhbhum Shear Zone, eastern India, using pyrite textures and compositions in the Turamdih U-Cu(-Fe) deposit. Miner Deposita 44, 61–80 (2009). [Google Scholar]
- Moreno L. & Neretnieks I. Long-term environmental impact of tailings deposits. Hydrometallurgy 83, 176–183 (2006). [Google Scholar]
- Li X., You F., Huang L., Strounina E. & Edraki M. Dynamics in leachate chemistry of Cu-Au tailings in response to biochar and woodchip amendments: a column leaching study. Environ Sci Eur 25, 32 (2013). [Google Scholar]
- Li X., You F., Bond P. L. & Huang L. Establishing microbial diversity and functions in weathered and neutral Cu–Pb–Zn tailings with native soil addition. Geoderma 247–248, 108–116 (2015). [Google Scholar]
- Ivanova J., Djingova R., Korhammer S. & Markert B. On the microwave digestion of soils and sediments for determination of lanthanides and some toxic and essential elements by inductively coupled plasma source mass spectrometry. Talanta 54, 567–574 (2001). [DOI] [PubMed] [Google Scholar]
- Li X., Huang L., Bond P. L., Lu Y. & Vink S. Bacterial diversity in response to direct revegetation in the Pb–Zn–Cu tailings under subtropical and semi-arid conditions. Ecol Eng 68, 233–240 (2014). [Google Scholar]
- Cayford B. I., Dennis P. G., Keller J., Tyson G. W. & Bond P. L. High-throughput amplicon sequencing reveals distinct communities within a corroding concrete sewer system. Appl Environ Microbiol 78, 7160–7162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imelfort M. & Dennis P. Ace Pyrotag Pipeline - wrapper for QIIME and R - interfaces with PyroDB (https://github.com/Ecogenomics/APP) (2011) (04/04/2014).
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg L., Stone G., Imelfort M., Hugenholtz P. & Tyson G. W. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9, 425–426 (2012). [DOI] [PubMed] [Google Scholar]
- Wu S. T., Zhu Z. W., Fu L. M., Niu B. F. & Li W. Z. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genomics 12, 444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imelfort M. & Dennis P. Normaliser (https://github.com/minillinim/Normaliser) (2011) (08/11/2013).
- Kolde R. A grid based implementation of heatmaps that offers more control over heatmap dimensions and appearance (http://CRAN.R-project.org/package=pheatmap) (2012) (10/03/2015).
- Pruesse E., Peplies J. & Glockner F. O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci 14, 927–930 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.