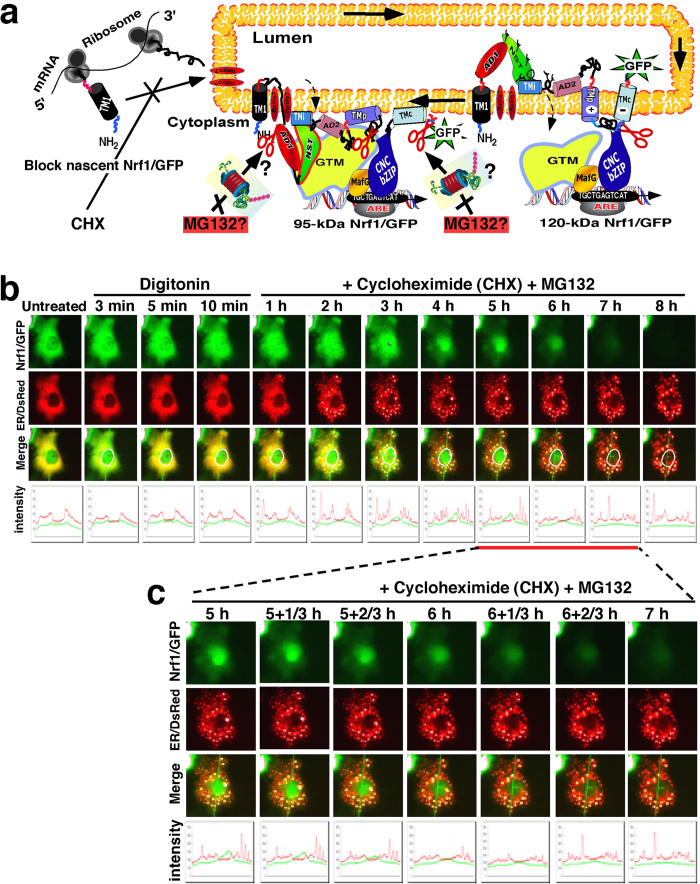
Figure 11. Live-cell imaging of the sensitivity of pre-existing Nrf1/GFP to the proteasome.
(a) Distinct membrane-topologies of the 120-kDa and 95-kDa Nrf1 proteins around and within the ER are schematically shown. Nrf1 has been fused C-terminally by GFP, which faces the luminal side of membranes during the initial topogenesis. Since Nrf1 is a mobile membrane-protein that entails dynamic topologies, the membrane-topological folding of glycosylated 120-kDa Nrf1/GFP is distinct from that of non-glycosylated/de-glyosylated 95-kDa Nrf1/GFP (also see refs 25,26). Once the C-terminal region of Nrf1 would be repartitioned across the ER membrane into the cyto/nucleoplasmic side, whereupon it is susceptible to proteolytic degradation mediated by the ERAD-dependent proteasome or other cytosolic proteases. The nascent Nrf1 protein from ribosomes can be blocked by cycloheximde (CHX). (b) COS-1 cells co-expressing Nrf1/GFP and the ER/DsRed marker were subjected to live-cell imaging combined with the chase experiment. The cells were permeabilized by digitonin (20 μg/ml) for 10 min, before being co-incubated with CHX (50 μg/ml) and MG132 (5 μmol/l) for 8 h. During the course of the experiment, real-time images were acquired using the Leica DMI-6000 microscopy system. The merged images of Nrf1/GFP with ER/DsRed are placed (on the third row of panels), whereas changes in the intensity of their signals are shown graphically (bottom). The features of arrow-indicated cells are described in the main text. Overall, the images shown are a representative of at least three independent experiments undertaken on separate occasions that were each performed in triplicate (n = 9). (c) Additional live-cell imaging of Nrf1/GFP was acquired from 5 to 7 h after co-incubation of CHX and MG132 in the presence of digitonin, as described above.