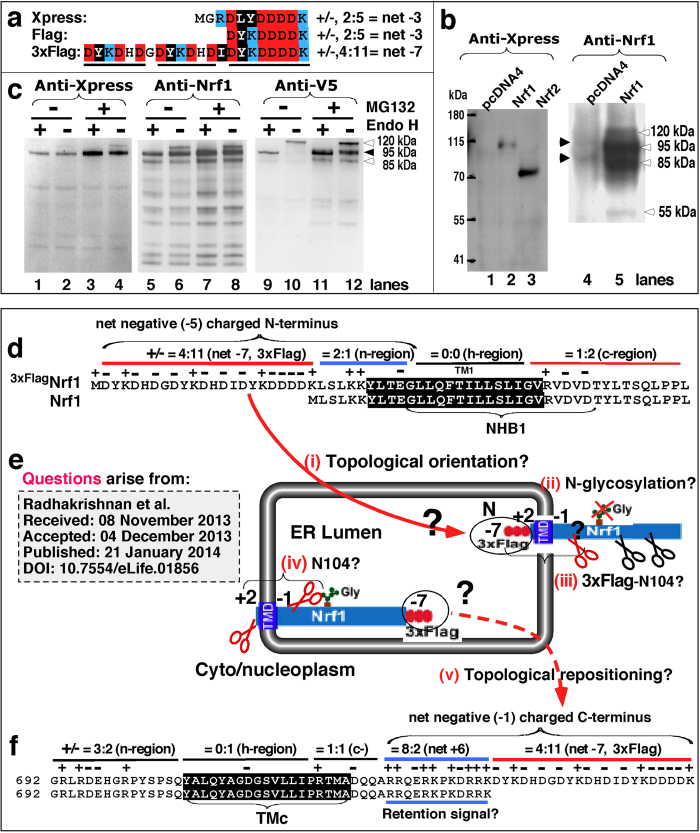
Figure 13. The membrane-topology of ectopic Nrf1 can be dictated by the negative charged 3xFlag or Xpress epitopes.
(a) Amino acid alignment reveals that the net negative charged Xpress epitope is highly conserved with Flag and 3xFlag. Acidic and basic amino acids are placed in the red and blue backgrounds, respectively, whereas the hydrophobic amino acids are on a black background. (b) Total lysates from COS-1 cells that had been transfected with pcDNA4/HisMax-based expression constructs for Nrf1, Nrf2, or an empty vector (i.e. pcDNA4), were separated by 7% NuPAGE in a Tris-Acetate running buffer, followed by immunoblotting with an Xpress antibody. The immunoblotted nitrocellulose membrane was then re-probed with an additional antibody against Nrf1 (right panel). (c) Total lysates from COS-1 cells expressing N-terminally Xpress-tagged Nrf1 or C-terminally V5-tagged Nrf1, which had been treated for 4 h with proteasomal inhibitor MG132, were subjected to Endo H-catalyzed deglycosylation reactions, before being resolved using 4–12% LDS/NuPAGE in Tris-Bis running buffer, and then immunoblotted using Xpress or V5 antibodies. The anti-Xpress blotted nitrocellulose membrane was re-probed with Nrf1 antibody (middle panel). (d) Amino acid alignment of the TM1-associated regions from 3xFlagNrf1 and the intact Nrf1. The charge difference between the 3xFlagged n-region (net -5) and the c-region (net -1) directs the TM1 region of 3xFlagNrf1 to adopt an Nlum/Ccyt orientation within the ER membrane. By contrast, the net positive n-region (+2) enables the TM1 region within intact Nrf1 to adopt an Ncyt/Clum orientation. (e) Several questions about Nrf1 processing that have arisen from recently published work40 have been re-interpreted in the main text, using current knowledge of membrane-topology55,56 as a guiding principle. Notably, the strongly negative charged 3xFlag dictates the folding of 3xFlagNrf1 and Nrf13xFlag to adopt non-native membrane-topologies, which make distinctive contributions to the post-translational processing of Nrf1. (f) Amino acid alignment of the TMc-associated regions from Nrf13xFlag and the intact Nrf1. The original positive (+6) C-terminus of Nrf1 enables it to be topologically repositioned into the cytoplasmic side of membranes, whereas the net negative (-1) 3xFlagged C-terminus of Nrf13xFlag favours location with the ER lumen.