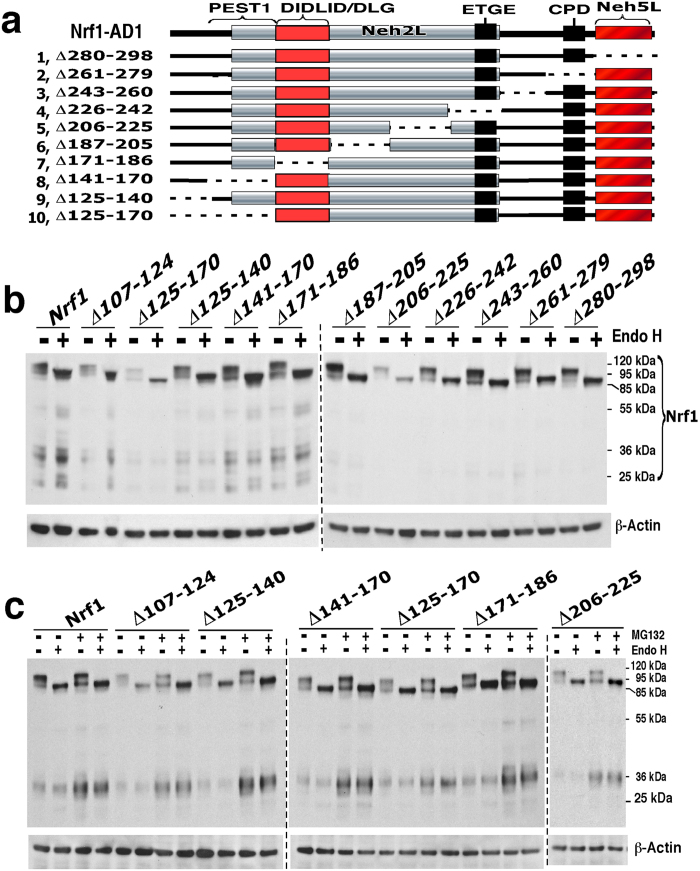
Figure 4. AD1 is required for Nrf1 stability and its glycosylation.
(a) Discrete regions within AD1, including PEST1, Neh2L (containing the DIDLID/DLG element and ETGE motif), CPD (i.e. 267LLSPLLT273) and Neh5L, are shown (upper cartoon). A series of progressive deletions (shown by lower hatched lines) were created from within the AD1 (aa 125–298) of Nrf1. (b) COS-1 cells were transfected with an expression construct for Nrf1 or the indicated mutants. Total cell lysates were subjected to deglycosylation reactions with Endo H. The reactants were resolved by 4–12% LDS/NuPAGE and visualized by immunoblotting. Removal of PEST1 and its adjoining portions within Neh2L gave rise to unstable proteins when compared to wild-type Nrf1. (c) Following overnight recovery from transfection with expression constructs for Nrf1 or mutants, COS-1 cells were treated with 5 μmol/l MG132 for 2 h before being harvested. Subsequent deglycosylation products were examined by immunoblotting. The presence of aa 107–170 and aa 206–225 appears to increase the basal abundance of Nrf1, and MG132 treatment did not obviously increase the levels of the 120-kDa mutant glycoprotein rather than the 95-kDa protein, indicating that the 120-kDa mutant proteins are subject to proteasome-independent proteolytic degradation.