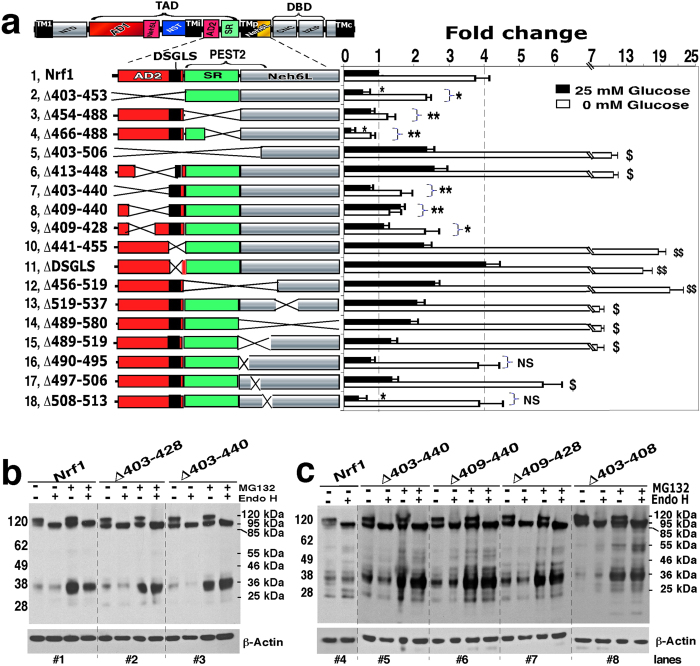
Figure 6. Differential regulation of Nrf1 by AD2, SR, PEST2, Neh6L and their adjacent degrons.
(a) The left schematic shows Nrf1 deletion mutants lacking various portions within AD2, including DSGLS-SDS1, SR, PEST2-SDS2 and Neh6L (Figs. S1D and S3A). The contributions of these regions to increased Nrf1 activity in response to glucose starvation (i.e. glucose-free medium), when compared with Nrf1 activity in medium fortified with 25 mM-glucose (control), were examined using an ARE-driven reporter assay. COS-1 cells that had been cotransfected with one of the expression constructs (1.2 μg), together with PSV40GSTA2-6 × ARE-Luc (0.6 μg) and β-gal plasmid (0.2 μg), were allowed to recover from transfection in fresh 5.5 mM-glucose medium for 8 h, before being cultured in no-glucose medium for a further 18 h. Transactivation of ARE-driven luciferase gene by Nrf1 and its mutants was determined as described in the text. The data were calculated as a fold change (mean ± S.D) of transactivation mediated by wild-type Nrf1 under 25 mM-glucose control conditions. Significant increases ($p < 0.05 and $$p < 0.001, n = 9) and decreases (*p < 0.05, **p < 0.001, n = 9) are indicated. (b and c) Cells expressing wild-type Nrf1, or mutants lacking the indicated portions of AD2, were allowed to recover from transfection for 24 h in 25-mM glucose-containing medium before treatment with 5 μmol/l MG132 for an additional 2 h. Following deglycosylation by Endo H, the proteins were resolved using LDS/NuPAGE containing 7% polyacromide gel in the pH 8.3 Tris-Acetate-SDS buffer and visualized by immunobloting with V5 antibodies. The amount of protein applied to each polyacrylamide-gel sample well was adjusted to ensure equal loading of β-gal activity. As indicated, the masses of the Nrf1 isoforms was estimated from their electrophoretic mobilities to be 120, 95, 85, 55, 46, 36 and 25 kDa, and β-actin was employed as an internal control.