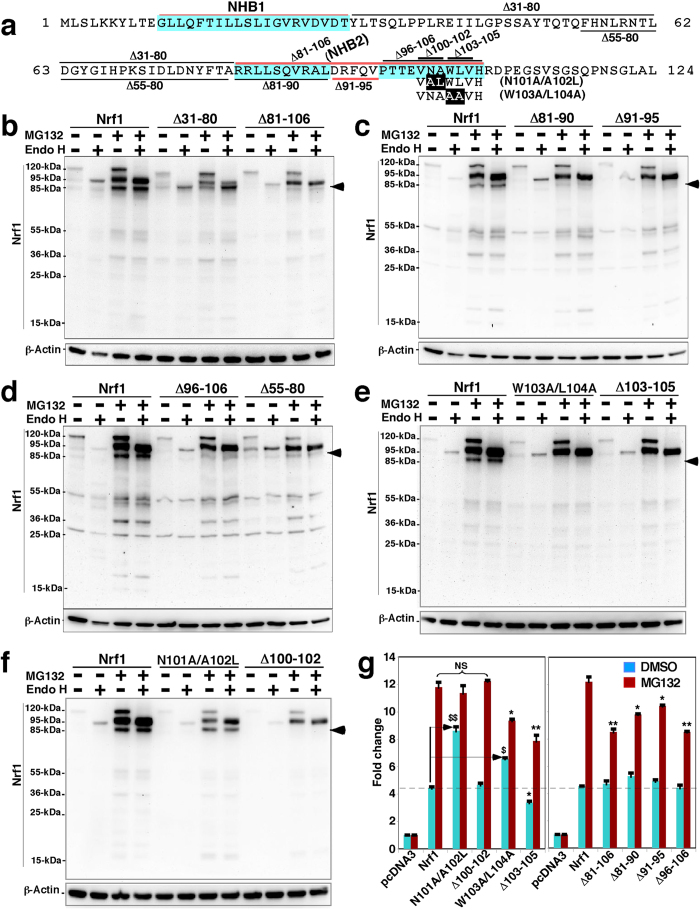
Figure 9. The abundance of cleaved 85-kDa Nrf1 protein is diminished by mutation of discrete regions around NHB2.
(a) Schematic representation of discrete regions, including NHB1 (aa 11–30) and NHB2 (aa 81–106), as well as several mutants (made herein), within the N-terminal domain (NTD, aa 1–124) of Nrf1. (b to f) Each expression construct for wild-type Nrf1, or the indicated NTD deletion mutants, was transfected into COS-1 cells for 6 h. Thereafter, the cells were allowed to recover from transfection for 24 h in 25-mM glucose-containing medium before being treated with 5 μmol/l MG132 for an additional 4 h prior to termination of the experiment. After in vitro deglycosylation reactions of denatured lysates with Endo H, the proteins were resolved using 4–12% LDS/NuPAGE in Tris-Bis running, and after transfer of the resolved proteins to membranes the blots were probed with antibodies against the V5 epitope. The masses of Nrf1 proteins based on their electrophoretic migration was estimated to be 120, 95, 85, 55, 46, 36 and 25 kDa. In addition, β-actin was employed as an internal control of protein loading. (g) After 6-h transfection with expression plasmids for Nrf1 or its mutants (1.0 μg), GSTA2-6 × ARE-Luc (0.5 μg), and pRL-TK (0.1 μg, used to control for transfection efficiency), the COS-1 cells were allowed to recover overnight in 25 mM-glucose medium before being treated with 1 μmol/l MG132 for 18 h, followed by measurement of ARE-driven transcription using the dual luciferase reporter assay. ARE-driven luciferase activity was normalized by the corresponding value of the Renialla activity, and then the results were calculated as a fold change (mean ± S.D) of Nrf1 mediated transactivation. Significant increases ($p < 0.05 and $$p < 0.001, n = 9) and decreases (*p < 0.05, **p < 0.001, n = 9) are indicated, relative to the background activity measured from pcDNA3-transfected cells (i.e. mediated by endogenous Nrf1 and/or Nrf2).