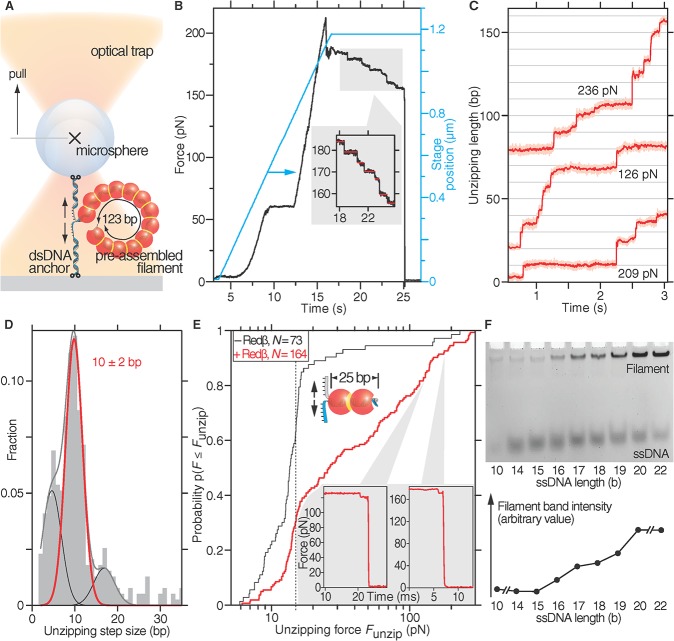
Fig 2. Nucleoprotein filaments consisting of at least a dimer resist substantial force.
(A) Schematic of high-force optical tweezers experiments to unzip preassembled nucleoprotein filaments (not drawn to scale). (B) A 123-bp nucleoprotein filament was subjected to increasing load forces (black line, left-hand axis) by moving the stage laterally (cyan line, right-hand axis). The inset shows a magnification of the stepwise dissociation after the stage movement was stopped. The horizontal red lines indicate steps. (C) Unzipping length as a function of time during the rupture of a 123-bp Redβ filament under constant force. Three typical traces are offset for clarity. (D) Unzipping step-size histogram of 391 Redβ dissociation events from (C). The step size was fitted as the sum of Gaussian curves (grey line) with the main peak (red line) occurring at 10 ± 2 bp and side peaks (black line) at about 5 bp and 17 bp. (E) Unzipping forces F unzip of a 25-bp region, preassembled with Redβ (red line, schematic) and without Redβ (black line), are plotted as a cumulative distribution function. The vertical dashed line indicates the average DNA unzipping force F unzip without Redβ. The insets show two examples of the stepwise drop in force prior to rupture taken from the indicated places. (F) Electrophoretic mobility shift assay to measure the minimum length required for a stable Redβ nucleoprotein filament. Complementary oligonucleotides of increasing lengths as indicated were incubated with a molar equivalent of Redβ before electrophoresis. The mean band intensities of the filament bands are plotted underneath the gel image.