Fig 5. Redβ oligomerizes with increasing concentration.
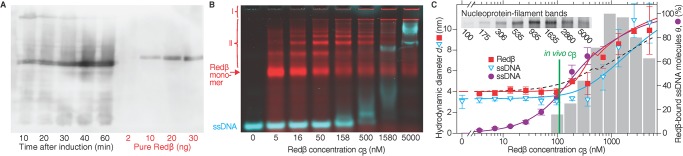
(A) Western blot after gel electrophoresis under denaturing conditions to determine the in vivo concentration of Redβ in E. coli induced to express at optimal conditions for Redβ-mediated homologous recombination. The band intensities were quantified and compared with purified StrepII-tagged Redβ (slightly slower migration due to the StrepII tag) loaded at the indicated concentrations. (B) Gel electrophoresis under nondenaturing conditions. Redβ banding pattern (red bands) indicates a concentration-dependent multimerization, which is not due to binding of ssDNA (cyan; 30 b, c ssDNA = 1 nM; see also Fig D in S1 Text). Below the gel pockets and stacking gel (I), bands due to oligomers and rings (II) are visible. (C) Hydrodynamic diameter (left axis) of Redβ (red squares), ssDNA (open cyan triangles), and their cross correlation (right axis, purple circles) based on fluorescence correlation spectroscopy (FCS) as a function of Redβ concentration c β averaged over three experiments. Error bars denote standard deviation. All plotted lines are calculated using our annealing model to compare the fit to the corresponding data points. The solid red line is based on the assumption of isodesmic growth with nucleation of a dimer (a dimer < a, reduced chi-squared value ), the dashed black line assumes isodesmic growth without nucleation (a dimer = a, ). The intensities of the inset nucleoprotein-filament gel bands obtained under nondenaturing conditions are shown as the grey histogram. The green line indicates the mean in vivo Redβ concentration of 110 nM.
