Abstract
Atherosclerosis has a high incidence and is harmful to human health. An elevated level of oxidized low-density lipoprotein (Ox-LDL) is one of the major risk factors for atherosclerosis. During atherogenesis progression, circulating monocytes adhere to the intima and differentiate into macrophages. After differentiation, intimal macrophages intake Ox-LDL via scavenger receptors, thereby transforming into foam cells. Foam cell formation due to excessive accumulation of cholesterol by macrophages is a pathological hallmark of atherosclerosis. To gain a molecular understanding of the effect of Ox-LDL in atherosclerosis development, we conducted a genome-wide analysis of the Ox-LDL-induced macrophage transformation by microarray gene expression profiling. Here we describe in details the contents and quality controls for the gene expression and related results associated with the data uploaded to Gene Expression Omnibus (accession number GSE54039).
Keywords: Ox-LDL, Foam cell, LncRNA, Macrophages, Microarray
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Strain(s) | Human monocytic THP-1 cells (Manassas, VA) |
| Sequencer or array type | Human LncRNA Array v2.0 (8 × 60 K, Arraystar) |
| Data format | Raw data: TXT files |
| Experimental factors | THP-1 macrophage cells without or with Ox-LDL treatment |
| Experimental features | Microarray gene expression profiling to identify genes that are regulated by Ox-LDL |
| Consent | N/A |
| Sample source | Location N/A |
Direct link to deposited data
Deposited data are available here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54039.
Experimental design, materials and methods
Cell line
The Human monocytic THP-1 cells were obtained from ATCC (Manassas, VA). It has been proved previously that THP-1 cells can differentiate into macrophages following treatment with 100 nM phorbol 12-myristate 13-acetate (PMA) for 72 h and macrophages can transform into foam cells by incubation in the presence of 50 μg/ml oxidized low-density lipoprotein (Ox-LDL) for 48 h. Therefore, THP-1 macrophage-derived foam cells have been used as a model system for investigating the possible molecular mechanisms that Ox-LDL influences foam cell development, which represents a promising therapeutic strategy for atherosclerosis.
Ox-LDL was obtained from Biomedical Technologies Inc. (Stoughton, MA). Accumulated evidence showed that Ox-LDL played a significant role in the initiation and progression of atherosclerosis [1]. The previous study revealed that an elevated level of Ox-LDL was one of the major risk factors for atherosclerosis. Circulating monocytes could adhere to the intima and then differentiate into macrophages [2]. After differentiation, intimal macrophages intake Ox-LDL via scavenger receptors, thereby transforming into foam cells [3]. To gain a molecular understanding of the mechanism by which Ox-LDL transforms the macrophages into foam cells, we generated THP-1 macrophage-derived foam cells by Ox-LDL treatment.
Long noncoding RNAs (lncRNAs) are nonprotein coding transcribed RNA molecules consisting of more than 200 nucleotides [4]. An increasing number of studies have revealed that lncRNAs have a variety of important functions, including their role in transcription, splicing, translation, nuclear factor trafficking, imprinting, genome rearrangement, and chromatin modification [5], [6]. Studies have shown that aberrant lncRNA expression is associated with a variety of human diseases such as cardiovascular diseases and cancer [7], [8]. Thus, a better understanding of the roles of lncRNA in atherosclerosis will advance us to find a promising strategy for regulating lipid metabolism balance.
Microarray and quality control
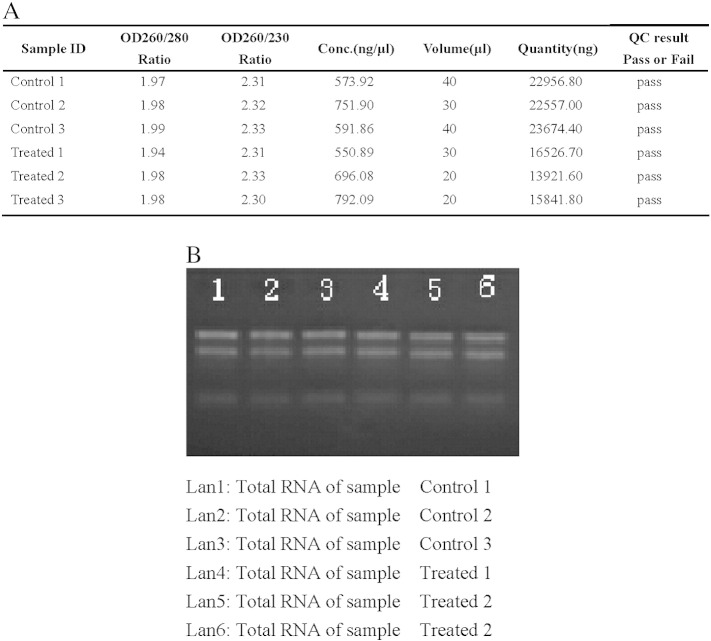
To identify the genes that are regulated by Ox-LDL, we isolated total RNA from 6 independent samples of THP-1 macrophages cultured in the presence or absence of 50 μg/ml of Ox-LDL in serum-free RPMI 1640 medium containing 0.3% BSA for 48 h using Trizol (Invitrogen Life Technologies). RNA from 6 samples was subjected to microarray analysis. Total RNA from each sample was quantified by the NanoDrop ND-1000 and RNA integrity was assessed by standard denaturing agarose gel electrophoresis (Fig. 1A and B). For microarray analysis, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols with minor modifications. Briefly, mRNA was purified from 1 μg total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Human LncRNA Array v2.0 (8 × 60 K, Arraystar). After having washed the slides, the arrays were scanned by the Agilent Scanner G2505B. Agilent Feature Extraction software (version 10.7.3.1) was used to analyze acquired array images. All microarray data were submitted to the Gene Expression Omnibus (accession number GSE54039).
Fig. 1.
Quality control of RNA. (A) RNA from 6 independent samples was quantified by NanoDrop ND-1000. (B) RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Control 1 to control 3 represent RNA from THP-1 macrophages (three samples). Treated 1 to treated 3 represent RNA from THP-1 macrophage-derived foam cells (three samples).
Normalization
Agilent Feature Extraction software (version 10.7.3.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, lncRNAs and mRNAs that at least 1 out of 6 samples have flags in Present or Marginal (All Target Values) were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs with statistical significance were identified through Volcano Plot filtering. Pathway analysis and GO analysis were applied to determine the roles of these differentially expressed mRNAs played in these biological pathways or GO terms. Finally, Hierarchical Clustering was performed to show the distinguishable lncRNA and mRNA expression pattern among samples.
Basic analysis
To explore possible changes in RNA expression during macrophage formation, we performed microarray analysis of THP-1 macrophages and THP-1 macrophage-derived foam cells using the Arraystar probe dataset, which included 24,748 lncRNAs and 24,420 coding transcripts. LncRNA and mRNA expression profiles from THP-1 macrophages (3 samples) and THP-1 macrophage-derived foam cells (3 samples) were produced using the Arraystar Human LncRNA Array v2.0 platform. Expression values were normalized based on the mean expression value for each probe set. Scatter plots of normalized signal values in lncRNAs and mRNAs are shown in Fig. 2A and B. Differently expressed probe sets were identified based on Student's t-test for paired samples' normalized expression values using the following cutoff: absolute fold change value larger than 3 and P value less than 0.01. By using a ≥ 3 fold change as a cut-off, P-value ≤ 0.01, 100 lncRNAs and 63 mRNAs were differentially expressed between THP-1 macrophage versus THP-1 macrophage-derived foam cells. Value distribution of signals from microarrays is shown in Fig. 3A and B.
Fig. 2.
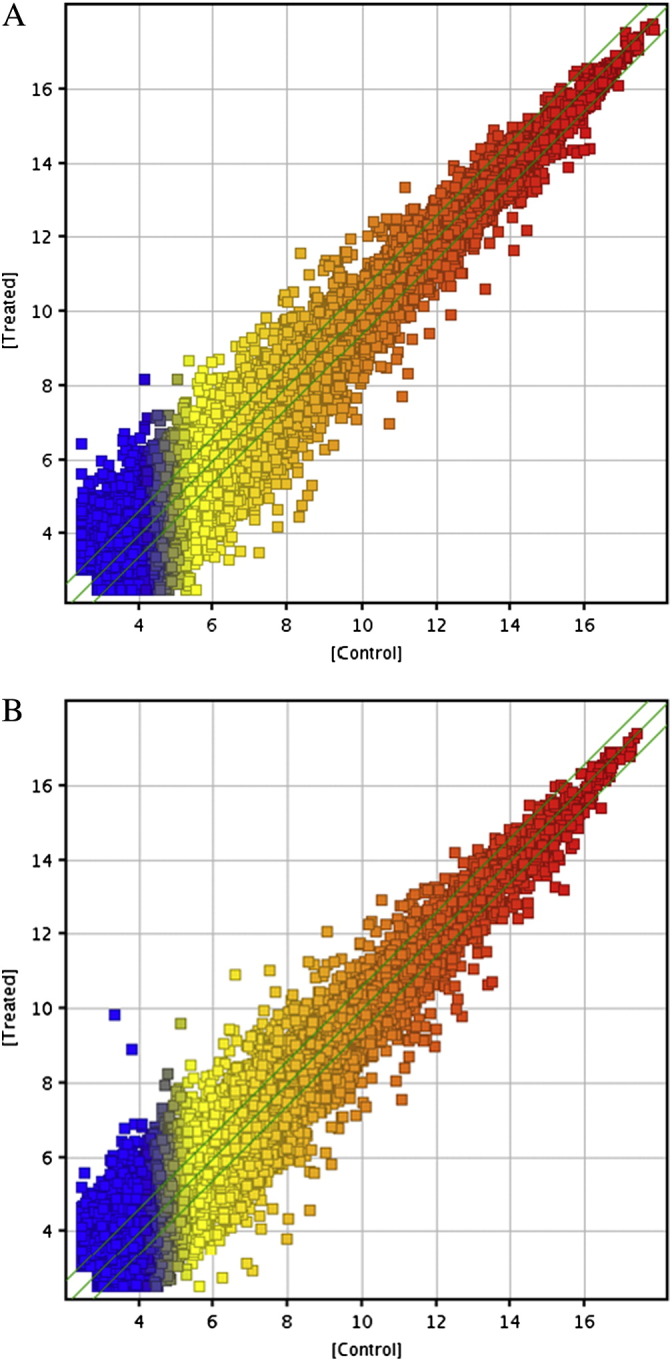
Scatter plots showing correlation of signal values between Control and Treated. (A) The lncRNA expression of Control (horizontal axis) vs Treated (vertical axis). (B) The mRNA expression of Control group (horizontal axis) vs Treated group (vertical axis). Control group represents THP-1 macrophages (three samples). Treated group represents THP-1 macrophage-derived foam cells (three samples). The values of X and Y axes in the scatter-plot are the normalized signal values of the samples (log2 scaled) or the averaged normalized signal values of groups of samples (log2 scaled). The green lines are Fold Change Lines (the default fold change value given is 1.5). The lncRNAs/mRNA above the top green line and below the bottom green line indicated more than 1.5 fold change of lncRNAs/mRNA between the two compared samples or the two compared groups of samples.
Fig. 3.
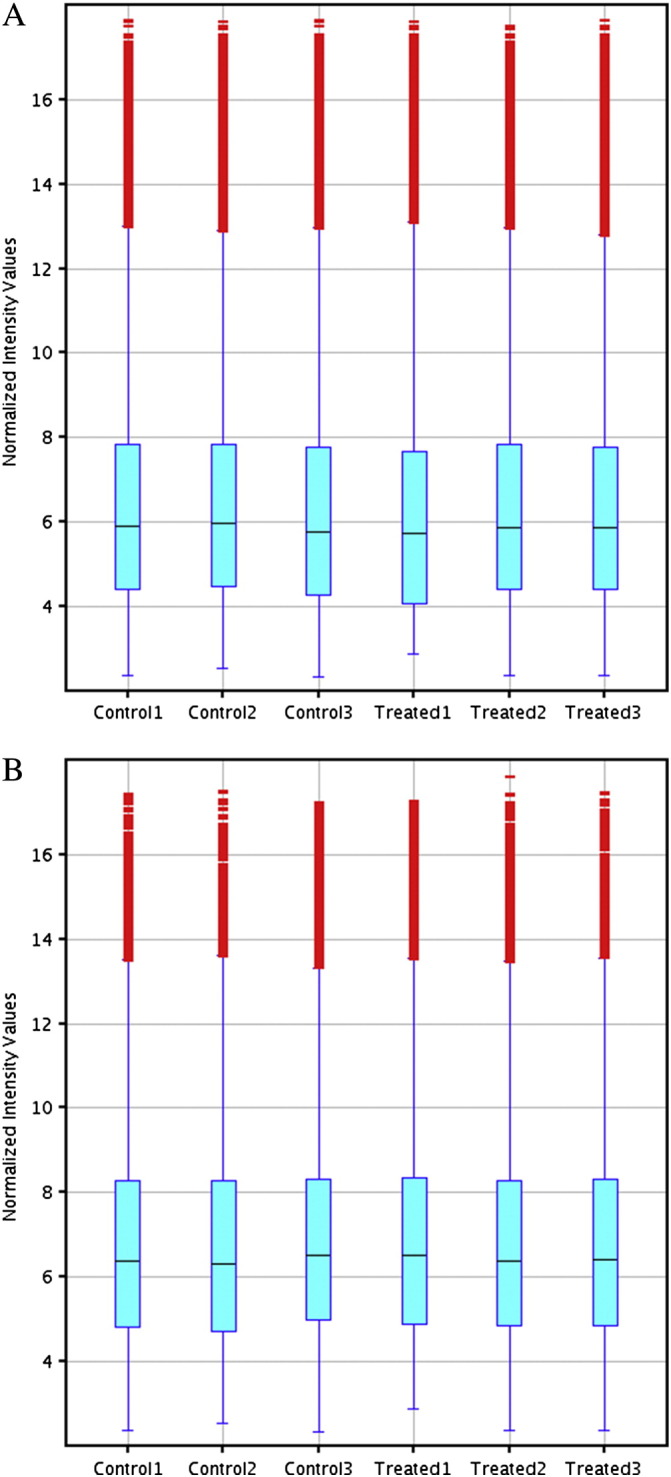
Value distribution of signals from microarrays. (A) indicates lncRNAs and (B) indicates mRNAs. The boxes mark the interval between the 25th and 75th percentiles. The lines inside the boxes denote medians.
Discussion
We described here a dataset composed of microarray expression profiling of Ox-LDL-responsive genes. With this dataset, we were able to demonstrate a large number of Ox-LDL-responsive lncRNA and mRNA, which advance us to further investigate their relationships and functions in foam cell formation and atherosclerosis progression. We believe that this dataset would be particularly valuable for investigating the cellular processes associated with lipid metabolism balance and the underlying molecular mechanisms. For example, we revealed that Ox-LDL could significantly promote lincRNA-DYNLRB2-2 expression, which upregulated ATP-binding cassette transporter A1 (ABCA1) expression through G protein-coupled receptor 119 (GPR119). In addition, lincRNA-DYNLRB2-2 and GPR119 promoted ABCA1-mediated cholesterol efflux from THP-1 macrophage-derived foam cells, and inhibited inflammatory responses [9]. Moreover, we paid particular attention to RP5-833A20.1 as it is a key lncRNA in our study of regulation of cholesterol homeostasis and inflammatory reaction. We found that lncRNA RP5-833A20.1 was located in intron 2 of the nuclear factor I/A (NFIA) gene and had an opposite transcription direction to that of NFIA through the UCSC Genome Browser. In addition, we demonstrated that RP5-833A20.1 could inhibit NFIA expression by promoting miR-382-5p expression. This finding sheds new light on the interplay between lncRNA and atherosclerosis, and suggests that lncRNA may become a promising strategy for regulating lipid metabolism balance [10].
Conflict of interest
The authors have no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant number 81301489).
References
- 1.Mitra S., Goyal T., Mehta J.L. Oxidized LDL, LOX-1 and atherosclerosis. Cardiovasc. Drugs Ther. 2011;25:419–429. doi: 10.1007/s10557-011-6341-5. [DOI] [PubMed] [Google Scholar]
- 2.Tabas I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 2010;10:36–46. doi: 10.1038/nri2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vicca S., Hennequin C., Nguyen-Khoa T., Massy Z.A., Descamps-Latscha B. Caspase-dependent apoptosis in THP-1 cells exposed to oxidized low-density lipoproteins. Biochem. Biophys. Res. Commun. 2000;273:948–954. doi: 10.1006/bbrc.2000.3017. [DOI] [PubMed] [Google Scholar]
- 4.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaral P.P., Dinger M.E., Mercer T.R., Mattick J.S. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., Chen Z., Wang X., Huang Z., He Z. Long non-coding RNA: a new player in cancer. J. Hematol. Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheuermann J.C., Boyer L.A. Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. EMBO J. 2013;32:1805–1816. doi: 10.1038/emboj.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y.W., Yang J.Y., Ma X., Chen Z.P., Hu Y.R. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J. Lipid Res. 2014;55:681–697. doi: 10.1194/jlr.M044669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y.W., Zhao J.Y., Li S.F., Huang J.L., Qiu Y.R. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler. Thromb. Vasc. Biol. 2014 doi: 10.1161/ATVBAHA.114.304296. pii: ATVBAHA.114.304296 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]