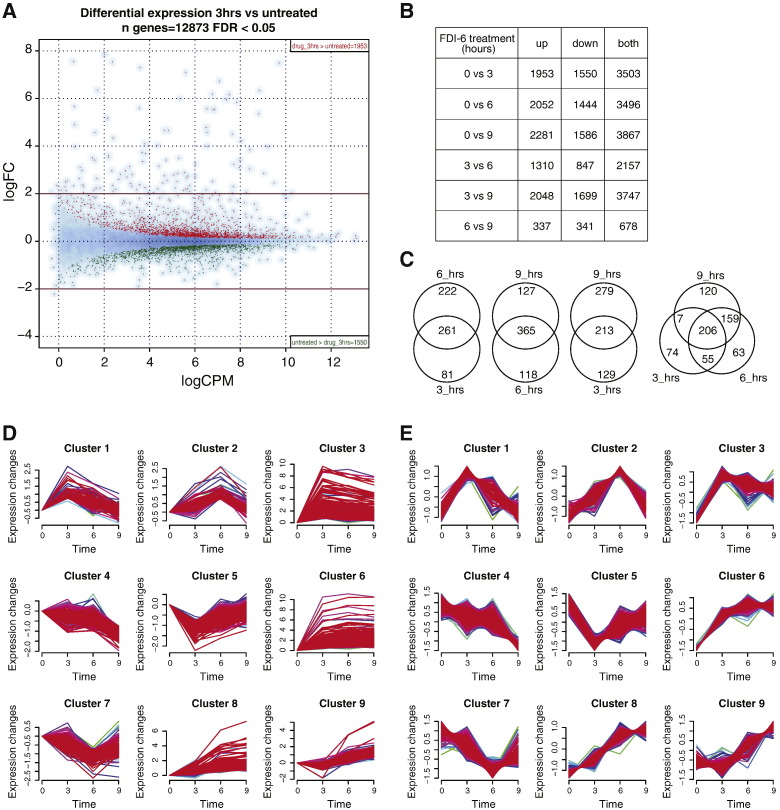
Fig. 2.
Differential expression and time clustering analysis. (A) Global differential expression map of RNA-Seq after 3 h treatment versus untreated. Y-axis: logarithm of fold change (logFC); X-axis: logarithm of counts per million of reads (logCPM). Red dots: up-regulated genes (n = 1951); green dots: down-regulated genes (n = 1552). FDR = false discovery rate. (B) Number of genes differentially expressed for each couple of time points. Up = upregulated in second time points compared to first one; down = downregulated in second time point compared to first one; both = some of up and down categories. Abbreviations as in Fig. 1C. (C) Venn diagrams showing the overlap of differentially expressed genes at the different time points. hrs = hours. (D) Temporal cluster analysis grouping genes that show similar changes in expression after FDI-6 treatment, original profiles. (E) Same as D, standardized profiles.