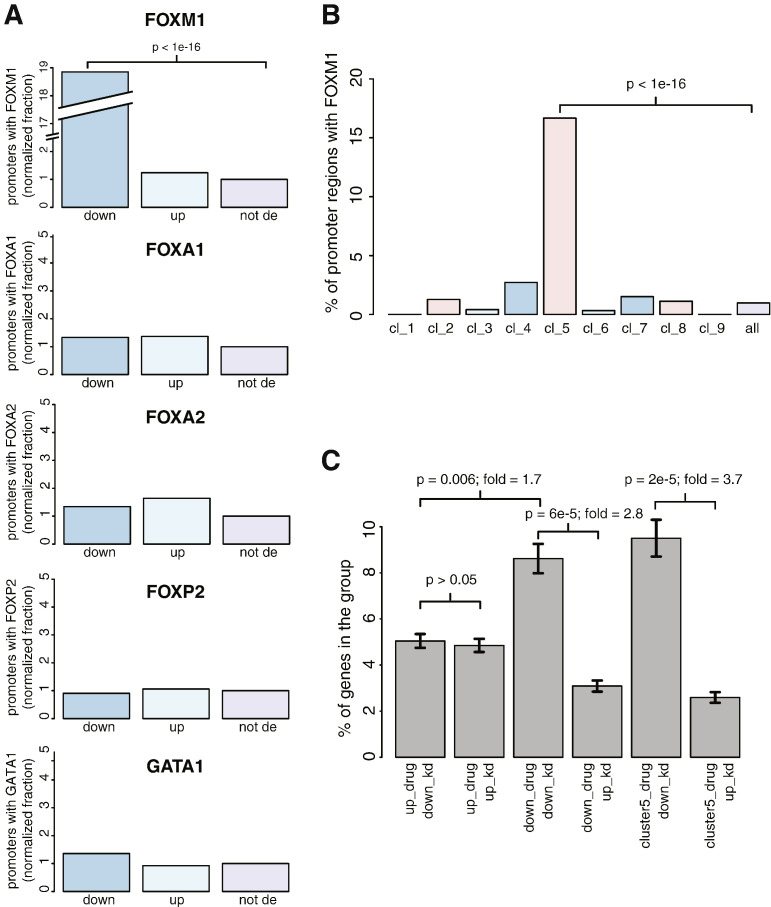
Fig. 3.
Cross validation with independent data sets. (A) Barplots showing the percentage of differentially expressed gene sets with a transcription factor occupancy in the promoter region (2 kb upstream of tss). Plots are shown for the closely related forkhead transcription factors FOXM1, FOXA1, FOXA2, FOXP2 as well as the transcriptional activator GATA1. For each plot, from left to right, bars represent down-regulated (down), up-regulated (up), and not differentially expressed (not de) gene sets. (B) Barplot showing the percentage of genes in the different temporal clusters with a FOXM1 peak in the promoter region. (C) Barplot showing the fraction of genes that were up- and down-regulated by FDI-6 and were commonly differentially expressed in an existing microarray data set of FOXM1 siRNA knockdown. Error bars indicate s.e.m.