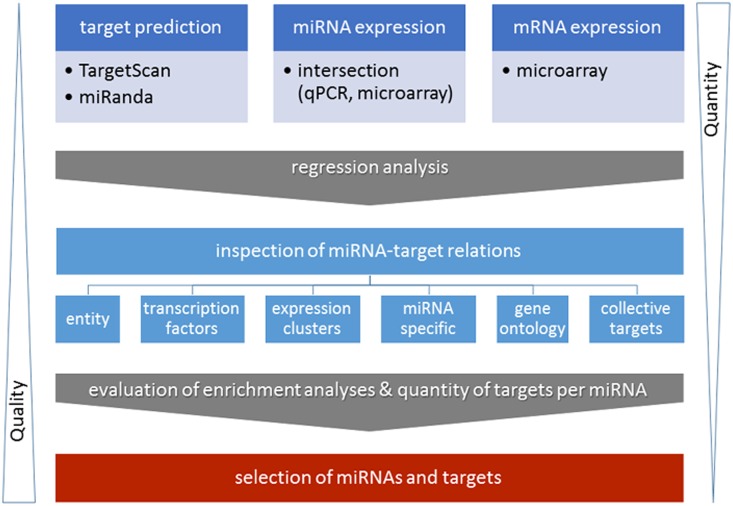
Fig 1. Joint analysis of miRNA and mRNA expression data.
Schematic overview of the integrative analysis of miRNA target prediction based on TargetScan (www.targetscan.org/) and miRanda (www.microrna.org/) and paired miRNA/mRNA expression data derived from the same experiments. The graphic is an extension and modification of Fig 1 published by Meyer and co-workers 2014 [15]. The expression data included the intersection dataset derived from miRNA microarray and miRNA qPCR profiling experiments and was inversely associated to mRNA microarray data. Predicted miRNA-target relations from integrated analysis were evaluated according to different criteria: Criteria which can be considered for mRNA-miRNA selection include: the total number of miRNA targets per miRNA, the number of transcription factors per miRNA, enrichment of targets in gene expression clusters, or gene ontology terms of all selected miRNAs or miRNA specific target enrichments, as well as the number of collective miRNA targets. The workflow allowed multi-aspect based interpretation of the results, reduced the quantity of miRNA-mRNA target relations, and increased the prediction quality with respect to potential biological implications.