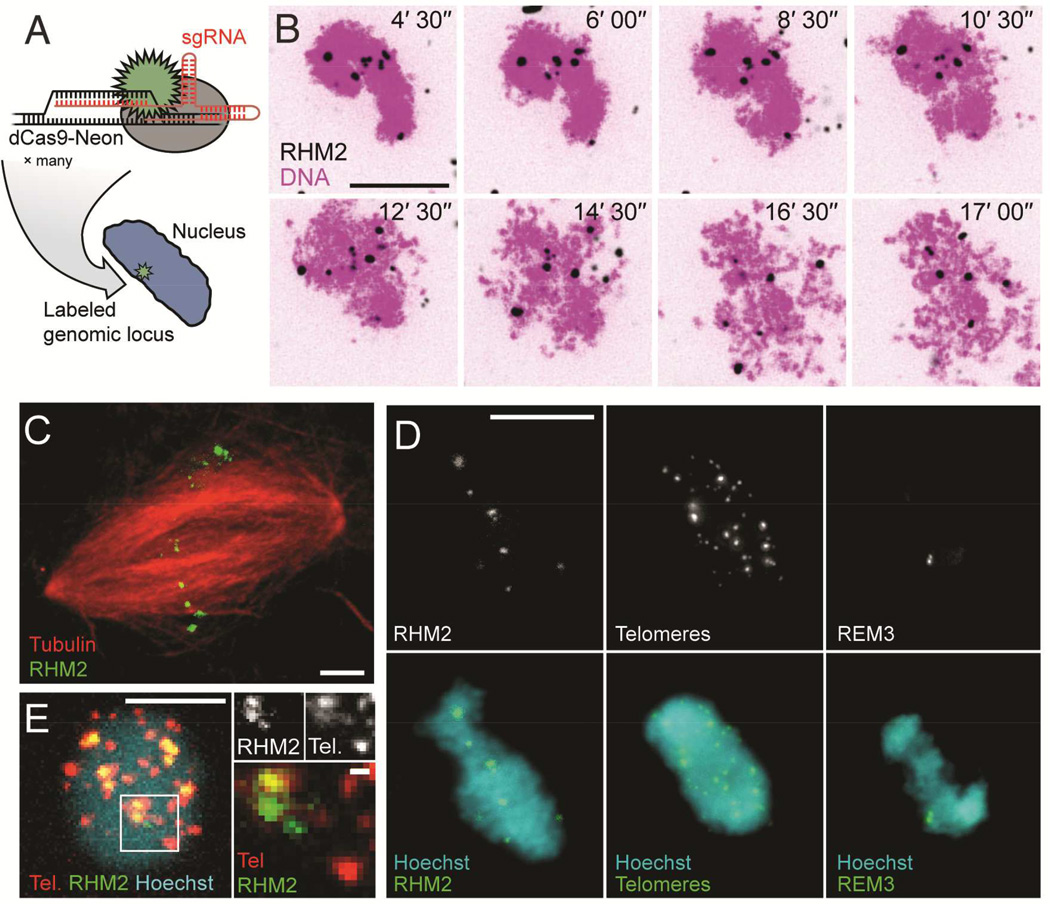
Figure 1. Repetitive genomic loci can be visualized using dCas9-Neon in Xenopus egg extracts.
A: dCas9-Neon is programmed to label specific genomic loci by conjugation to an sgRNA molecule containing a complementary target sequence. See also Figure S1. B: dCas9-Neon programmed using RHM2 sgRNA (black) localizes rapidly to loci in sperm nuclei (Sytox Orange dye, magenta). Time (min) after imaging started is indicated in the top left of each image. See also Supplementary Movie 1 and Figure S2. C: Labeled RHM2 loci (green) are maintained following formation of a mitotic spindle (red). D: Three examples of repeat classes labeled on sperm nuclei in Xenopus egg extract (1n = 18). Left: RHM2 is a centromere-proximal locus on ~65% of chromosomes (Freeman and Rayburn, 2005). Middle: Telomere repeats target chromosome termini. Right: REM3 is reported to target a single centromere-proximal locus on chromosome 1, appearing here as two spots (Hummel et al., 1984). E: Left: Sperm nuclei driven into interphase in the presence of dCas9-tdTomato Telomere sgRNA and dCas9-Neon RHM2 sgRNA demonstrate simultaneous dual-color labeling (scale bar, 5 µm). Right: A subset of RHM2 and telomere loci appear to co-localize, while others do not (scale bars 10 µm, except magnification in panel E, 1 µm).