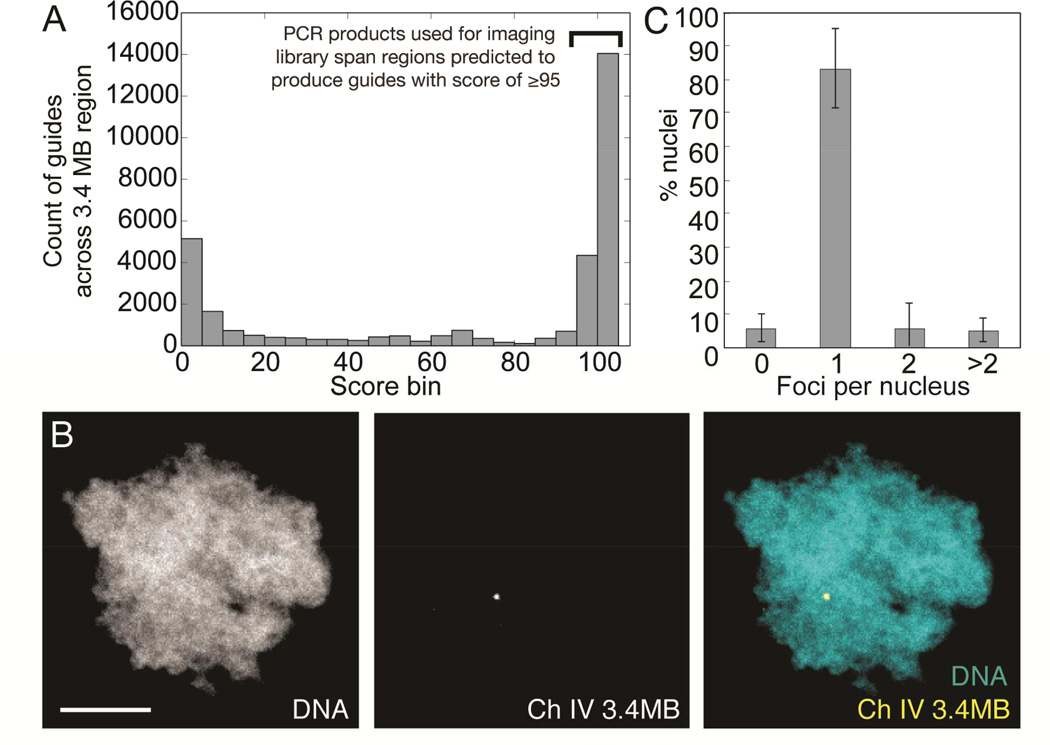
Figure 3. A single 3.4 MB locus can be labeled using an enzymatically generated guide library.
A: Specificity score distribution for all guides predicted to be generated by subjecting 3.4 MB region to procedure outlined in Figure 2A. Only sub-regions predicted to generate guides with a score of ≥95 were used as PCR templates for library construction. B: Processing of 100 PCR products (See Figure S3) spanning regions within a 3.4MB region of X. laevis chromosome 4 generates a single labeled spot in haploid sperm nuclei (scale bar, 5 µm). C: Count of fluorescent foci per sperm nucleus when incubated with 3.4 MB library. n = 3 experiments, 11–13 nuclei scored per experiment. Bars are ± standard deviation. See also Figure S3, Table S3, and Supplemental Data S1–S3.