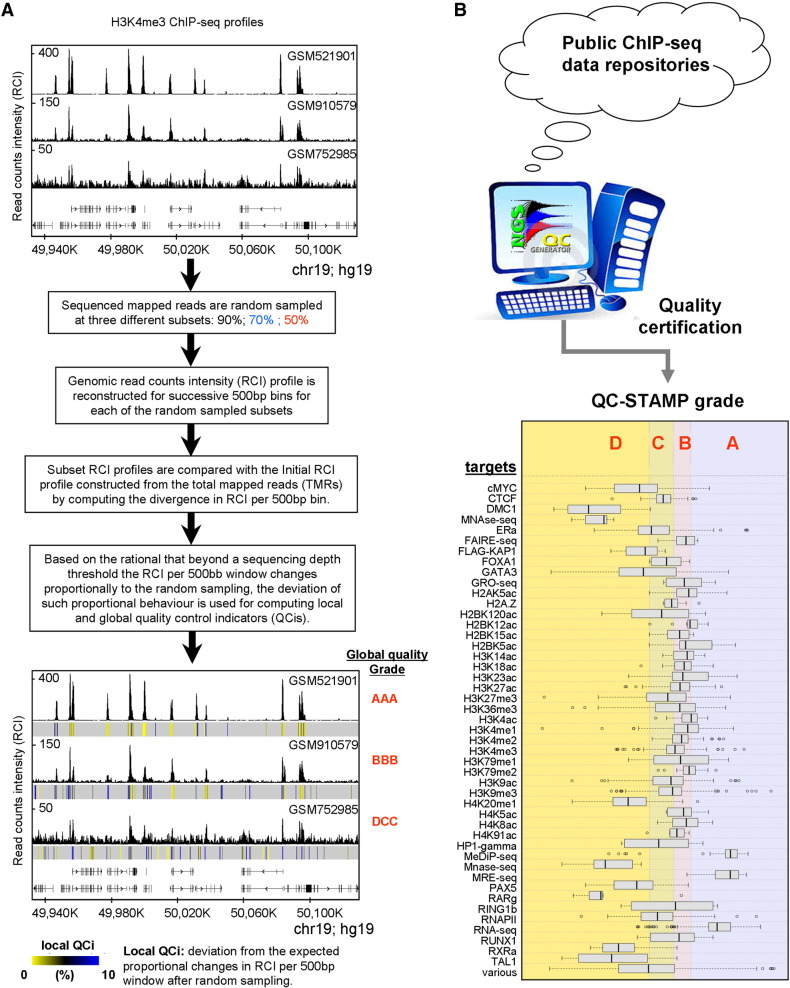
Fig. 1.
Strategy applied by the NGS-QC Generator for assessing quality descriptors for ChIP-seq profiles. (A) Genome-browser screenshot illustrating three publicly available H3K4me3 ChIP-seq datasets. Based on a visual inspection we could conclude that, while all three profiles share common binding sites, important differences in their read count intensity levels as well as in their background noise are observed. These datasets were subjected to the NGS-QC Generator pipeline for assessing quality descriptors. Briefly, TMRs are randomly sampled into three distinct populations (90, 70 and 50%), which are used for profile reconstruction by computing the RCIs in 500-bp bins. The RCI divergence from expectation is measured relative to the original profile (s100). This information generates local quality indicators (QCis) and is displayed together with the original RCI profile to identify robust chromatin regions (local QCi heat-map below the bottom profiles). In addition, global quality descriptors are computed which are summarized into the Global quality Grade or QC-STAMP. In this particular example, the best H3K4me3 dataset received a “AAA” grade while the worst has been discerned a “DCC” QC-STAMP. It is worth to mention that the intuitive quality assessment performed by the visual inspection is now comforted by a global and quantitative QC descriptor. (B) The NGS-QC Generator has been used to perform quality certification of datasets retrieved in publicly available repositories. Currently, more than 8000 datasets were certified covering a variety of data types and were classified based on a quality score grades from AAA for the highest quality datasets to DDD for those presenting the worst, like input control datasets.