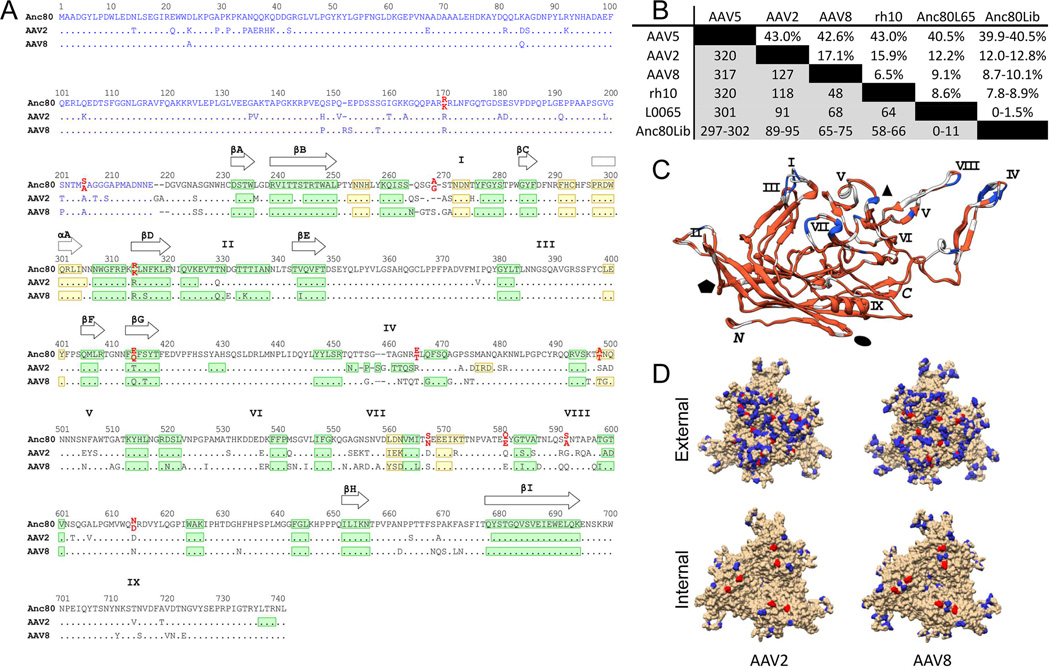
Figure 2. Sequence and structural analysis of Anc80 vectors.
A. Sequence alignment of Anc80, AAV2 and AAV8 VP3 proteins. A structural alignment derived from the crystal structures of AAV2 (PDB 1LP3) and AAV8 (PDB 2QA0) VP3 and the predicted structure of Anc80L65 VP3 was generated with UCSF Chimera (Pettersen et al., 2004) and is represented in black print. The blue region is a non-structural alignment of the VP1/VP2 domains of AAV2, AAV8 and An80 (Notredame et al., 2000). Ambiguous residues in Anc80Lib are in red print with the lower position corresponding to Anc80L65 residues. β-strands and α-helices are represented in green and yellow, respectively. The positions of the nine β-strands forming the AAV antiparallel β-barrel are depicted with plain arrows whereas the position of the conserved core α-helix is depicted with a dotted arrow. The approximate positions of variable regions (VR) I-IX are represented by the roman numerals above the sequence alignment. B. AAV Cap Sequence divergence matrix: Table represents above the diagonal the percent sequence divergence from selected AAV serotypes, as well as rh.10, most homologous VP1 sequence as by BLAST. Below the diagonal, the number of amino-acid differences per position is presented. C. Superimposition of AAV2 and AAV8 VP3 crystal structures with Anc80L65 VP3 predicted structure. The color code depicts the amino acid conservation between the 3 aligned sequences of panel A (red: highest conservation; blue: lowest conservation). Variables regions I-IX and C/M-termini are indicated in black. The approximate positions of the two, three and five-fold axis are represented by the black ellipse, triangle and pentagon, respectively. D. Structural mapping of amino-acid changes as compared to AAV2 (left) and AAV8 (right) on VP1 trimer visualizing the external (top) and internal (bottom) of the virion. Colored residues are divergent in Anc80. Red colored residues are ambiguous via ASR and therefore dimorphic in Anc80Lib.