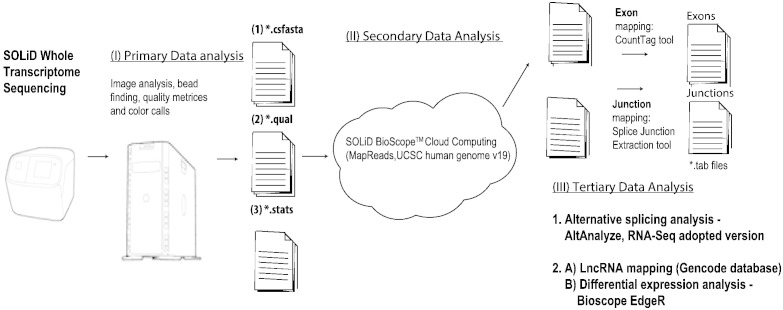
Fig. 2.
Whole transcriptome sequencing on the produced libraries yielded .csfasta, .qual and .stat files for each sequenced sample. Mapping to the human genome (UCSC database) was conducted through cloud computing to produce both exon and junction level quantification files. AltAnalyze software was used to analyze these files separately and combined and detect high confidence splice isoform alteration between the different clinical states. Long noncoding RNAs (lncRNAs) were detected in the sequenced libraries through mapping to the GENCODE catalog (version 7), and differential expression analysis conducted for the lncRNAs using the Bioconductor project EdgeR program.