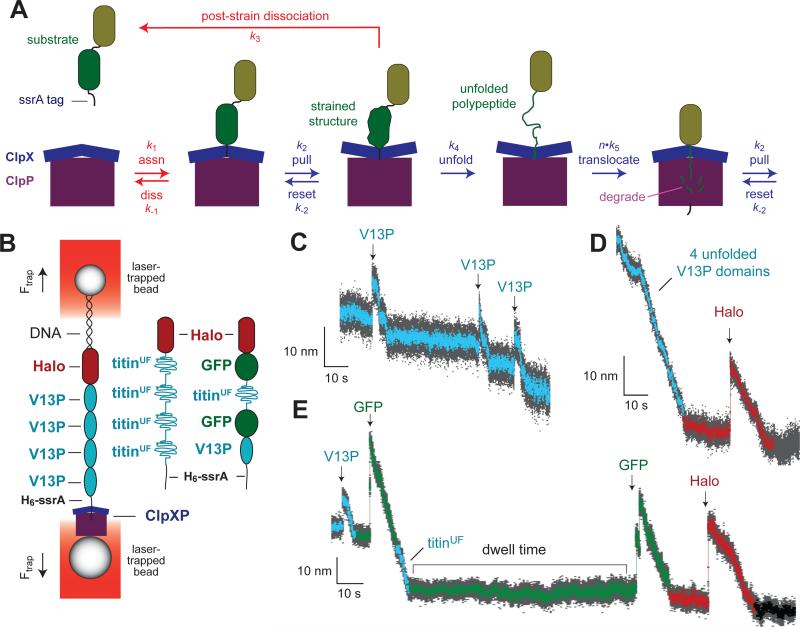
Figure 1.
ClpXP degradation. (A) Model for ClpXP degradation of a two-domain protein bearing an ssrA degradation tag. Blue arrows are kinetic steps that contribute to unfolding and translocation in solution degradation and in optical-trap experiments, whereas red arrows are steps that only contribute to degradation in ensemble assays. The k1 association step requires ATP binding but not hydrolysis. Mechanical steps (k2 and k5) are power strokes fueled by ATP hydrolysis. In an optical-trap experiment, dissociation by the k−1 or k3 pathways would terminate the experiment. Similarly, following degradation of the green domain, release of the olive domain by a k3-like step would end an optical-trap experiment. (B) Cartoon of optical-trap experiment. ClpXP bound to one laser-trapped bead engages a multi-domain substrate bound to a second laser-trapped bead, creating a stretched tether that offsets a force that would otherwise move the beads apart as a consequence of the laser positions and power. The beads move farther apart when ClpXP unfolds a native domain and closer together when ClpXP translocates an unfolded segment of the substrate. These changes in bead-to-bead distance provide an assay of the mechanical activities of ClpXP. Different substrates used in optical-trap studies are depicted schematically. (C) Optical-trap trace of AAAYYY ClpXP degradation of the Halo-(V13P)4-H6-ssrA substrate, showing three V13P unfolding events (arrows) and subsequent translocation at an experimental force of 5.2 pN. In this and subsequent panels, bead-to-bead distances decimated to 300 Hz are gray and those decimated to 30 Hz are colored. (D) Trace of AAYYYY ClpXP degradation of the Halo-(V13PCM)4-H6-ssrA substrate at 7 pN. Cyan symbols correspond to translocation of the unfolded V13P titin domains, whereas dark-red symbols show the pre-unfolding dwell time, unfolding (arrow), and translocation of the Halo domain. (E) Trace of AYYYYY ClpXP degradation of the Halo-GFP-titinUF-GFP-V13P-ssrA substrate (8 pN). Cyan symbols represent titin events, green symbols represent GFP events, and dark-red symbols represent Halo events.