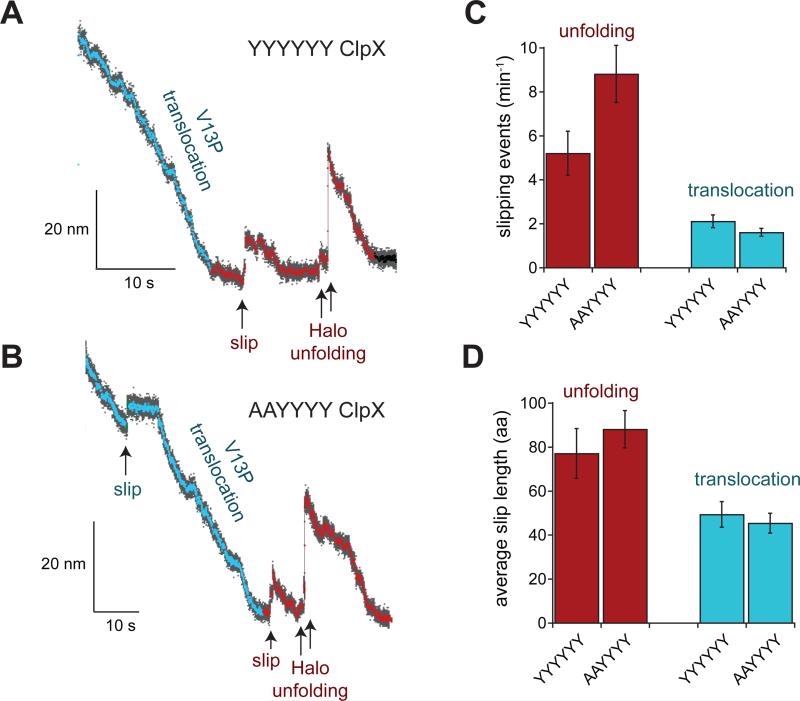
Figure 4.
Slipping during translocation and unfolding of the Halo-(V13PCM)4-H6-ssrA substrate by the YYYYYY and AAYYYY ClpX enzymes in the absence of ClpP. Data were recorded at average forces of ~10 pN (range 6-14 pN) for both YYYYYY and AAYYYY. (A) Individual trace with a slipping event for YYYYYY ClpX during the pre-unfolding dwell in an optical-trap experiment at 6.5 pN. Bead-to-bead distances decimated to 300 Hz are shown in gray and to 30 Hz in color. In this trace, the Halo domain unfolds in two steps, probably corresponding to extraction of the C-terminal helix followed by unfolding of the remaining structure. Similar two-step Halo unfolding was observed in 70-80% of the traces with ClpX alone or with ClpXP. (B) Individual trace with slipping events during translocation and the pre-unfolding dwell by AAYYYY ClpX in an experiment at 12.5 pN. Decimation and coloring as in panel A. (C) Number of slipping events min−1 for YYYYYY and AAYYYY ClpX during the Halo pre-unfolding dwell and translocation. (D) Average slip length for YYYYYY and AAYYYY ClpX during the Halo pre-unfolding dwell and translocation. In panels C and D, values are the mean ± SD of averages calculated for 35 independent trials in which 33% of the data was randomly discarded.