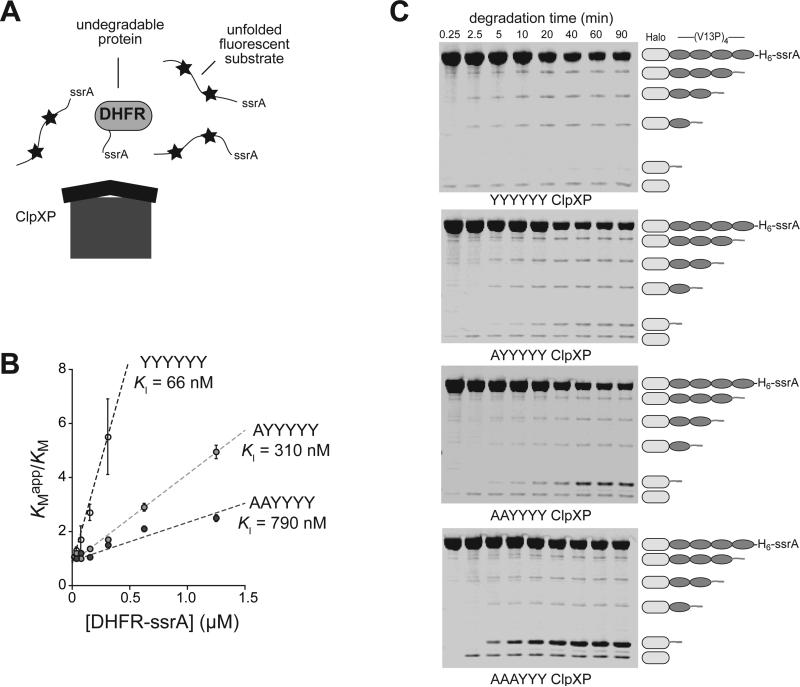
Figure 6.
Effects of p1-loop mutations on release of undegraded protein domains. (A) In the presence of methotrexate, DHFR-ssrA cannot be degraded by ClpXP and acts as a competitive inhibitor of the degradation of an unfolded ssrA-tagged substrate labeled with fluorescent dyes (stars), which are auto-quenched in the undegraded protein. (B) KMapp/KM for degradation of the fluorescent unfolded substrate by YYYYYY, AYYYYY, and AAYYYY ClpXP plotted against the concentration of the DHFR-ssrA competitor. Values are averages (N = 3) ± SEM. The lines are linear fits to KMapp/KM = 1 + [DHFR-ssrA]/KI. Degradation rates and apparent KM and Vmax values for different concentrations of the fluorescent substrate by ClpX variants (0.1 μM), ClpP (0.3 μM), and ATP (4 mM) were determined in the absence of competitor and in the presence of different concentrations of competitor (Fig. S3). (C) Degradation of TAMRA-Halo-(V13P)4-ssrA by the parental ClpXP and the p1-loop variants was assayed by SDS-PAGE. Fluorescent bands corresponding to TAMRA-containing fragments were detected using a Typhoon imager. Degradation reactions were performed at 30 °C and contained ClpX variants (1 μM hexamer), ClpP (2 μM tetradecamer), fluorescent substrate (10 μM), and ATP (10 mM). The lowest band on the gel was observed in a control experiment without ATP, suggesting that it arises from contaminating protease activity. Although larger fragments should appear faster in these experiments, those containing three, two, or one V13P domains were found to arise at similar rates. This behavior is expected for a reaction in which the rate of substrate engagement is slow compared to the rate at which individual V13P domains are degraded, especially if substrate release upon encounter with a V13P domain is rare.