Fig. 1.
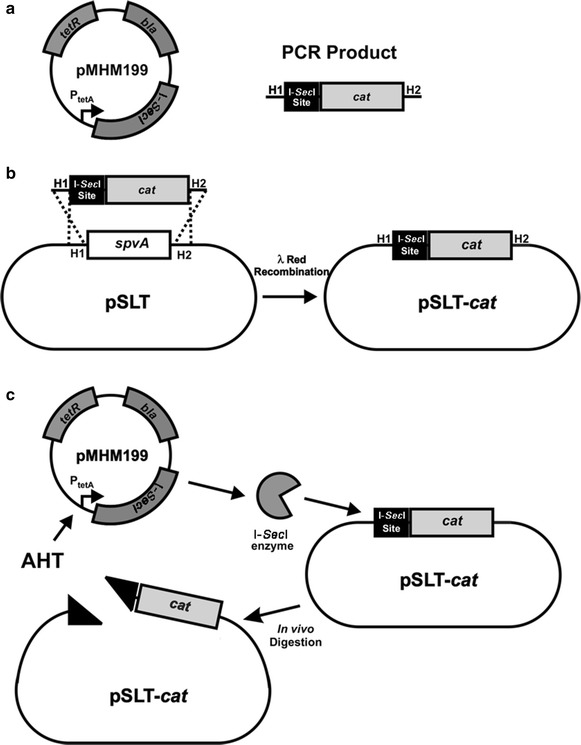
Steps involved in plasmid curing. a Plasmid pMHM199 contains I-SecI restriction endonuclease gene under the tetracycline inducible promoter PtetA, the regulator tetR, and the ampicillin resistance gene as well as the oriR101 origin of replication and temperature-sensitive repA101ts. b Deletion cassette is built by PCR using primers ATGAATATGAATCAGACCACCAGTCCGGCACTTTCACAGGTCGAAACCGCCGCCTTACGCCCCGCCCTGC and CTAAACTGCCGGCTGGCACGCAGAGTACCCGCAATCAACTGTTCCACCTGCTAGACTATATTACCCTGTT and plasmid pWRG100 as a template [in bold, regions of homology with pWRG100 (Blank et al. 2011)]. The cassette contains the I-SecI site adjacent to the chloramphenicol resistance gene cat, flanked by the homology regions to spvA gene (H1 and H2). c λ Red recombination step. The homology regions on the insertion cassette recombine with the spvA plasmid gene by λ Red recombination, excluding the spvA and substituting it with the I-SecI site linked to the cat gene generating the strain MHM21. d In vivo digestion. The plasmid pMHM199 is electroporated into MHM21, which is then plated onto LB agar containing ampicillin to select the positive transformants, and anhydrotetracycline.
