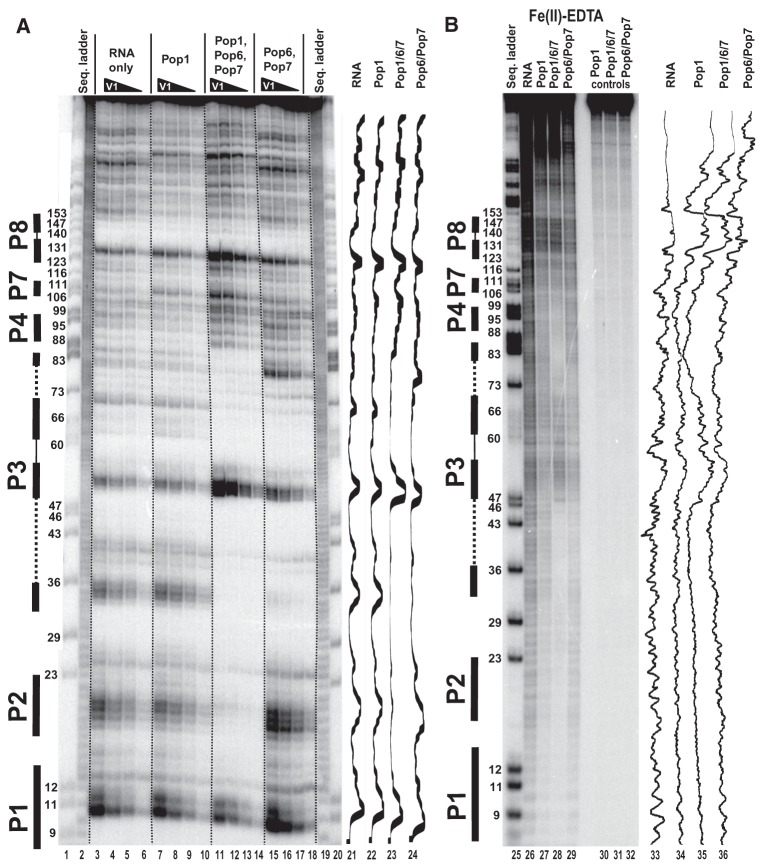
FIGURE 6.
Footprinting assays for RNase P RNA, typical gels. 5′-end 32P-labeled RNase P RNA alone or in complexes with equimolar amounts of Pop1 and the Pop6/Pop7 heterodimer (as indicated above the gels) was partially digested with RNase V1 (A) and hydroxyl radicals (B). (Lanes 1,20,25) RNase P RNA digested with RNase T1 (sequence ladder, identifies positions of guanines, shown on the left); (lanes 2,19) alkali hydrolysis of RNase P RNA (ladder); (lanes 3–6,26) digestion of RNase P RNA; (lanes 7–10,27) digestion of the complex of RNase P RNA with Pop1; (lanes 11–14,28) digestion of the quaternary complex of RNase P RNA with proteins Pop1, Pop6, and Pop7; (lanes 15–18,29) digestion of the complex of RNase P RNA with the Pop6/Pop7 heterodimer; (lanes 30–32) controls (RNA incubated with proteins as indicated above the gels). Secondary structure elements are marked on the left of the gels: Helical regions are shown by thick solid lines, terminal loops by thin solid lines, and the internal loop in the P3 RNA domain by dotted line. Gel traces are shown on the right of the gels (lanes 21–24,33–36) as marked on top.