FIGURE 2.
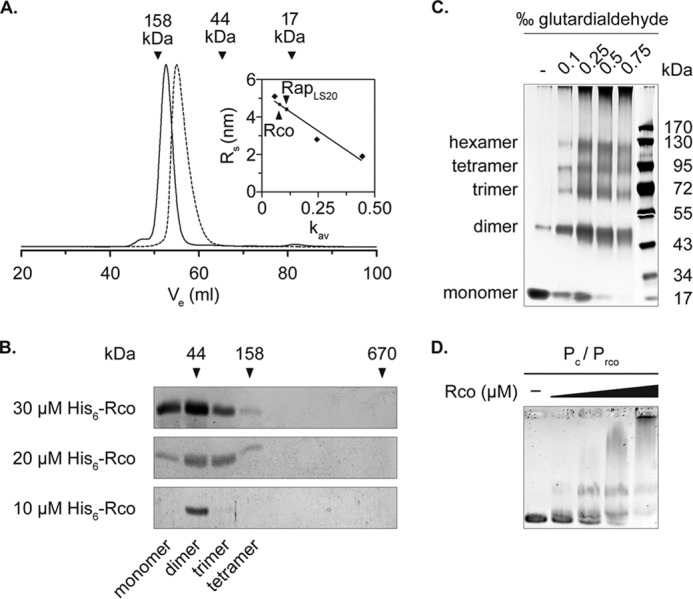
Rco and RapLS20 form dimers in solution. A, size exclusion chromatography of Rco and RapLS20. Vertical arrows, elution volumes of the standard proteins used to determine the molecular weight of Rco and RapLS20 (solid and dashed lines). The inset shows the standard curve used for calculating the Stokes radius of Rco (RS = 4.64) and RapLS20 (RS = 4.38). Size exclusion chromatography was performed on a HiLoad 16/600 Superdex 75 prep grade column (GE Healthcare). B, SDS-PAGE analysis of the 5–20% sucrose gradient sedimentation performed with different concentrations of Rco. Shown are fractions 1–9; arrows indicate the sedimentation behavior of the standards run in parallel. The molecular conformations of Rco below the SDS-PAGE analysis were calculated using the Monte-Siegel formula. C, chemical cross-linking of Rco with increasing concentrations of glutardialdehyde. Samples were run on 10% Tris-glycine PAGE (Nusep). D, binding of His6-Rco to the intergenic region of the repressor gene and the first gene of the conjugation operon containing the promoter of the conjugation operon (Pc) and the repressor gene (Prco). A 50 nm concentration of the DNA fragment was mixed with 0.2, 0.4, 0.8, and 1.6 μm Rco.
