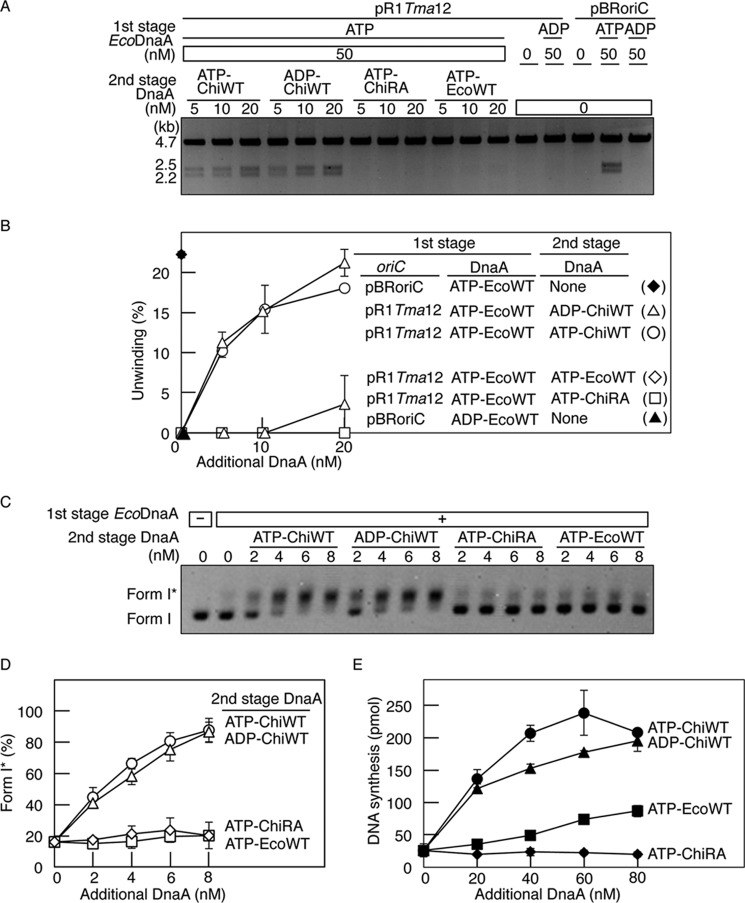
FIGURE 3.
Role for the Arg finger of R1 box-bound DnaA in initiation. A and B, DUE unwinding assay. In the first stage, pBRoriC (WT) or pR1Tma12 (4 nm as plasmid) was incubated on ice in buffer containing ATP-EcoDnaA (ATP-EcoWT) or ADP-EcoDnaA (ADP-EcoWT) (50 nm). In the second stage, the indicated amounts of ATP-chiDnaA (ATP-ChiWT), ADP-chiDnaA (ADP-ChiWT), ATP-chiDnaA R285A (ATP-ChiRA), or ATP-EcoDnaA were added to each mixture. Reactions were incubated at 38 °C for 3 min prior to digestion with P1 nuclease and AlwNI. Digestion products were analyzed by 1% agarose gel electrophoresis (A). The gel image is shown in a black-white inverted mode. The relative amounts of digested DNA to input DNA were quantified and used as a measure of DUE unwinding (%) (B). Two independent experiments were done, and both data and mean values are shown. C and D, form I* assay for DnaB helicase loading. The indicated amounts of DnaA proteins, as described above, were incubated at 30 °C for 20 min in buffer containing pR1Tma12 (1.6 nm as plasmid) and ATP-EcoDnaA (16 nm) in the presence of DnaB helicase, DnaC helicase-loader, SSB, and gyrase. Reactions were assessed using agarose gel electrophoresis analysis (C). The gel image is shown in a black-white inverted mode, and the migration positions of form I and form I* DNA are indicated. The amounts of form I* relative to total DNA were quantified and used as a measure of form I* (%) (D). Two independent experiments were done, and both data and mean values are shown. E, reconstituted replication assay. The indicated amounts of DnaA proteins, as described above, were incubated at 30 °C for 30 min in buffer containing pR1Tma12 (5.1 nm as plasmid; 600 pmol as nucleotides), ATP-EcoDnaA (60 nm), and replicative proteins. Two independent experiments were done, and both data and mean values are shown.