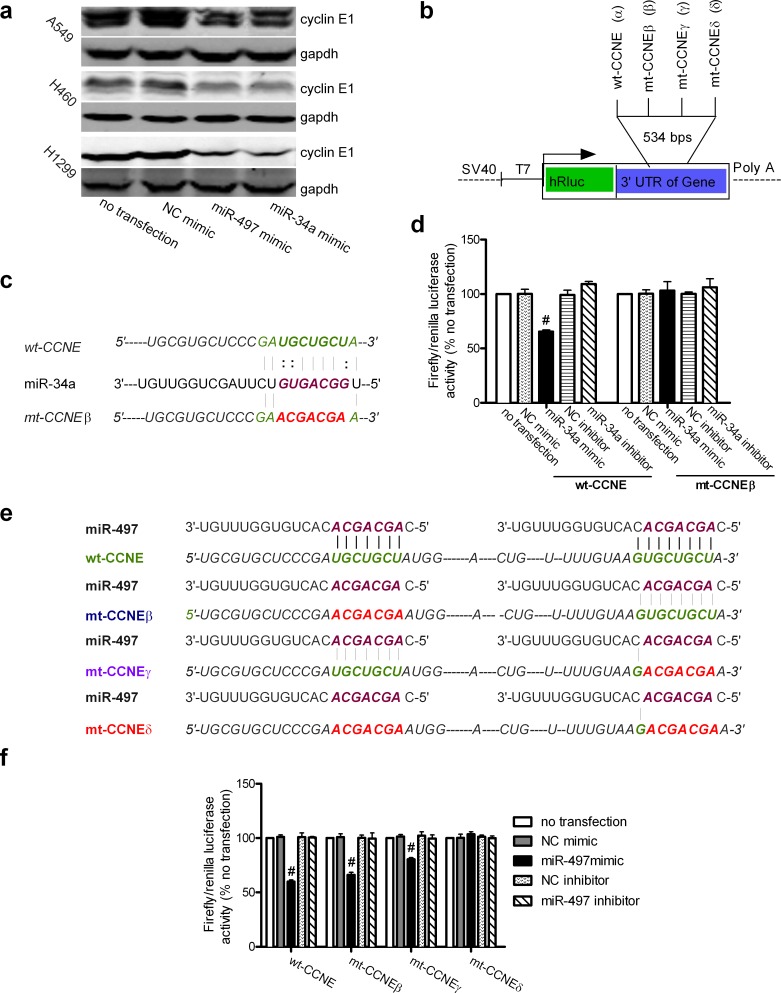
Figure 3. CCNE1 is a direct target of miR-497 and miR-34a.
(a) The expression levels of the cyclin E1 protein were measured by immunoblotting using GAPDH as the loading control. Experiments were performed in triplicate, with results similar to those shown. (b) Diagram of the psiCHECK2 vector used for the luciferase reporter activity assay. The arrow indicates the transcription start site. The magnified panel shows the location of the wild-type CCNE1 3′-UTR (α, wt-CCNE) or mutant CCNE1 3′-UTR at nt 247–253 (β, mt-CCNEβ), at nt 483–490 (γ, mt-CCNEγ), and at both (δ, mt-CCNEδ), which were subcloned into the psiCHECK2 vector. Thus, four individual plasmids were generated: wt-CCNE, mt-CCNEβ, mt-CCNEγ, and mt-CCNEδ. (c) Diagram of the wt-CCNE and mt-CCNEβ reporter constructs. mt-CCNEβ contains a seven-base mutation in the miR- 34a target region, abolishing its binding to miR-34a. (d) Firefly luciferase reporter activity assay. The relative luciferase activity was normalized to the Renilla luciferase activity and compared with that in the no-transfection control. Mean ± SD, n = 3 (#P < 0.01 vs. NC mimic). (e) Outline of the wt-CCNE, mt-CCNEβ, mt-CCNEγ, and mt-CCNEδ reporter constructs. mt-CCNEβ and mt-CCNEγ each contain one seven-base mutation in the miR-497 target region (at nt 247–253 or nt 483–490, respectively, in the CCNE1 3′-UTR), partly abolishing their binding to miR-497. mt-CCNEδ contains two seven-base mutations in the miR-497 target regions (at nt 247–253 and nt 483–490 of the CCNE1 3′-UTR), completely abolishing its binding to miR-497. (f) Firefly luciferase reporter activity assay. Relative luciferase activity was normalized to the Renilla luciferase activity and compared with that in the no-transfection control. Mean ± SD, n =3 (#P < 0.01, all vs. NC mimic).