Figure 1. Marked intraclonal variation in tumor-derived IGHV gene sequences in MM.
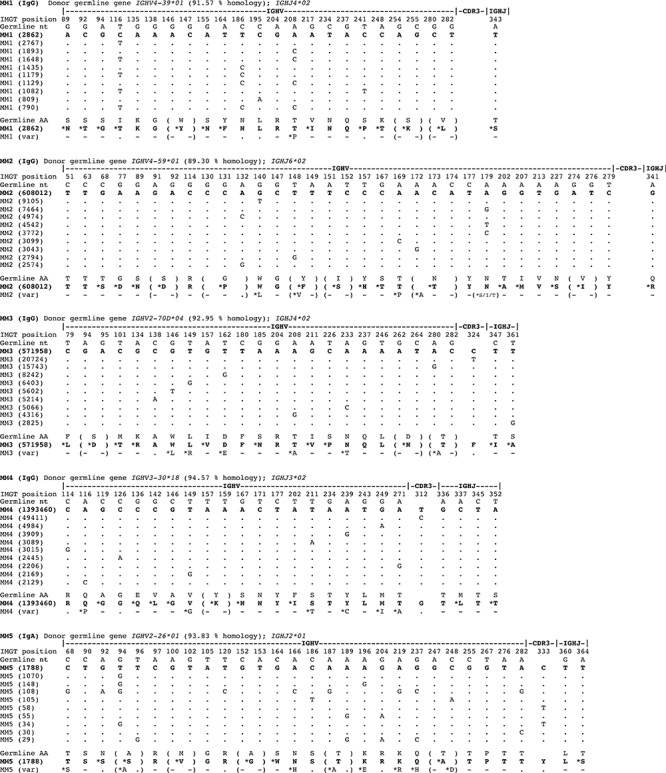
The pool of IgG transcripts identified from deep sequencing were aligned by computational analysis with donor germline IGHV genes and CDR3 identity used to delineate tumor-derived clonal sequences. Only selected informative codons are shown, focusing on revealing replacement nucleotides that arise by SHM for purposes of clarity. The % homology to germline IGHV gene of dominant clonal sequence in MM1-4 is stated in each case, and the dominant sequence is shown aligned to the germline gene. Data on the top 10 clonally-derived MM tumor IGHV sequences are shown aligned to the dominant sequence or top read, and not the germline gene to reveal the significant intraclonal variation observed in mutation patterns (dots denote sequence identity to the top read). For MM1 and MM2 aligned sequence matches start at position 43, for MM3 at position 65, and for MM4 at position 68. Translation to deduced amino acid (aa) sequences are shown aligning germline aa with replacement aa resulting from mutations in tumor IGHV gene sequences (denoted with *, and no change as -). No dominant clonal IGHV gene sequence was identified in the control MM5.
