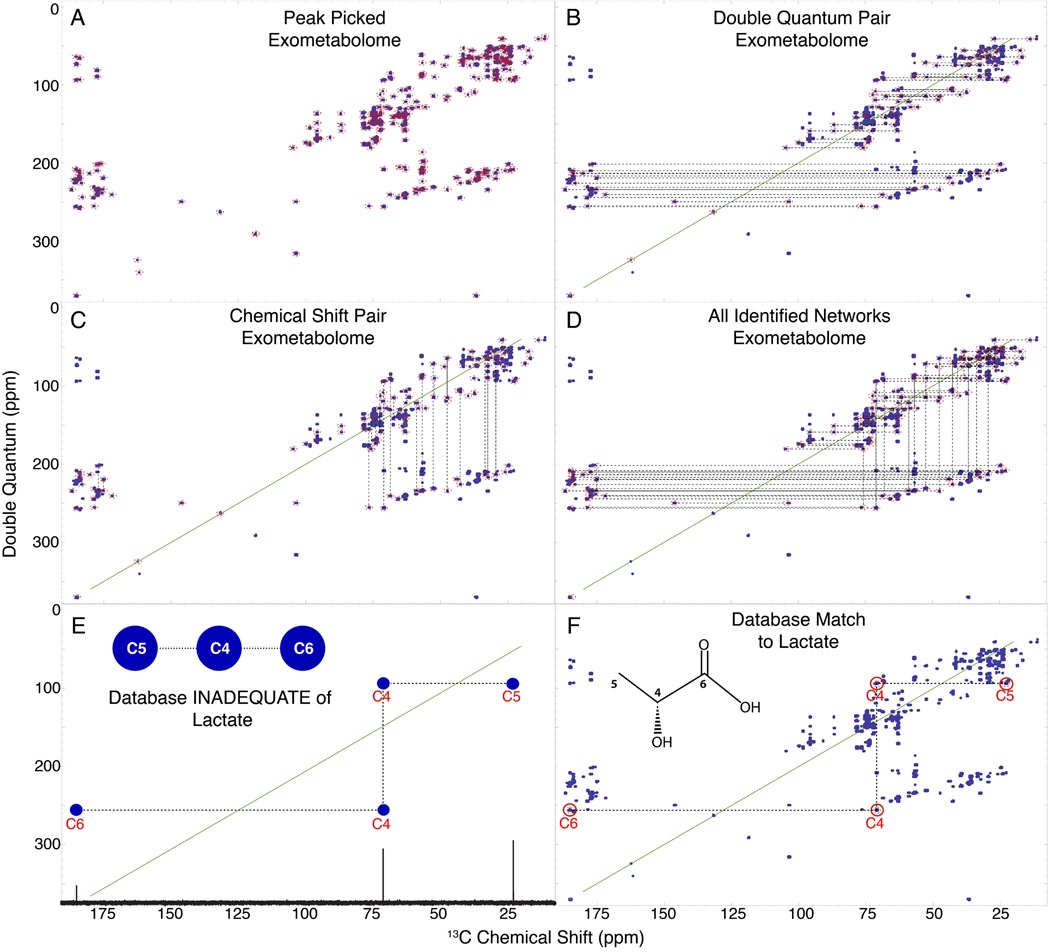
Figure 3.
INETA uses the rules of INADEQUATE to connect peaks. Peaks are picked (A) and their double quantum pair is found (B). Peaks lacking a partner are not considered in the analysis. Vertical pairs are matched based on chemical shifts (C). Vertical and horizontal pairs are connected to form a network. All networks are shown in D. 1D 13C spectra and carbon connectivity information is downloaded from the BMRB and a 2D INADEQUATE in silico database is generated by adding the chemical shifts of two directly bonded carbons (E). The INADEQUATE of database matches or candidates can then be plotted onto the peakpicked experimental spectra (denoted by red circles above carbon numbers) to give positive identification (F).