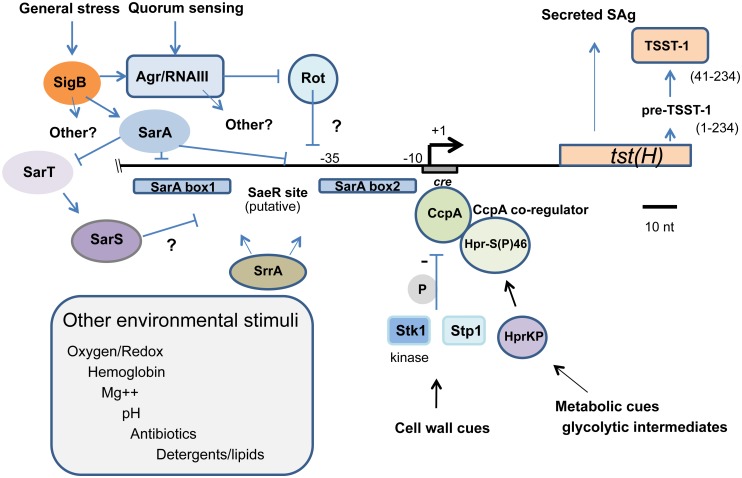
Fig 6. Model depicting the regulation of the TSST-1 superantigen in S. aureus.
The horizontal line shows the position of the transcriptional start site (+1) determined in RN4282 and 37 nucleotides upstream of the translation start site [40]. The positions of SarA boxes 1 and 2 are shown and correspond to their positions determined by both SarA consensus and direct DNA binding assay [40]. Negative regulators determined in the study reported herein include σ B, SarA, and Rot. SarA modulation of SarS is thought to occur via SarA negative regulation of the SarS activator SarT [65]. Catabolite control protein a (CcpA) binding to its cognate cis- acting cre site mediates additional tst repression by integrating signals from glycolytic intermediates via phosphorylation of the CcpA co-regulator as well as direct phosphorylation (grey P) via the Stk1 kinase which affects its DNA binding affinity [25, 44, 69]. The Stk1 S/T kinase and its cognate phosphatase, Stp1, may impart additional levels of control via the phosphorylation of SarA and the nucleoid protein, Hu, for example [45, 70, 71]. SrrA, the response regulator of the SrrAB two-component sensor, is thought to control tst regulation in response to oxygen and coenzyme Q [29, 30, 69, 100]. SrrA specific binding has been detected in the tst promoter region (Andrey, manuscript in preparation). DNA sequence with strong similarity to the consensus DNA binding site for the response regulator SaeR is shown. Although the precise involvement of SaeRS in tst regulation is unknown, it may help coordinate response to pH together with sigB [34, 101, 102]. Additional environmental stimuli known to affect TSST-1 expression are boxed although the precise genetic factors mediating these effects have yet to be defined [32, 102–107].