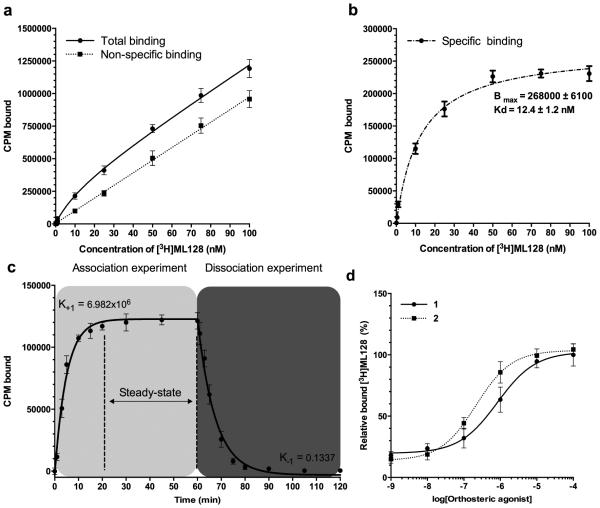
Figure 2.
Equilibrium analysis of compound 5 binding to mGlu4 in CHO cells revealed co-operative binding with orthosteric agonist. (a) Total and nonspecific binding were analyzed using 0.1, 0.5, 1.0, 10, 25, 50, 75, and 100 nM of ([3H]3). Method was based on whole cell binding assay, where incubations (30 min at room temperature) and washings (at +4 °C) were made in constant 100 nM concentration of glutamate. Sensitive centrifugation method was used to extract the unbound and bound fraction from each other. Nonradioactive 3 (10 μM, 1000x excess to hot ligand) was used to block specific binding site. (b) The specific binding was calculated based on the difference between analyzed total and nonspecific binding and presented as an average together with the standard deviation. The data was analyzed using GraphPad Prism software to estimate the presented Bmax and Kd values. (c) The association of the ([3H]3) was measured using 10 nM of hot ligand with or without 1000 fold excess of unlabeled compound to assess the nonspecific binding. Specific binding data are plotted at 0, 1, 3, 5, 10, 15, 20, 30, 45, and 60 min time points. The dissociation study was started 1 hour after the labeling of mGlu4 expressing cells with 10 nM of [3H]3 by adding (time = 60 min) 1000 fold excess of ([3H]3). Aliquots were removed at 61, 63, 65, 70, 75, 80, 90, 105 and 120 min time points, and the specific binding was determined. The K+1 and K−1 were analyzed using GraphPad Prism software and the estimated Kd was calculated as Kd= K−1/ K+1. (d) Co-operative binding between 1or 2 and ([3H]3) was studied by addition of orthosteric agonists. The specific binding is presented together with standard deviation from three experiments (n=3).