Figure 4.
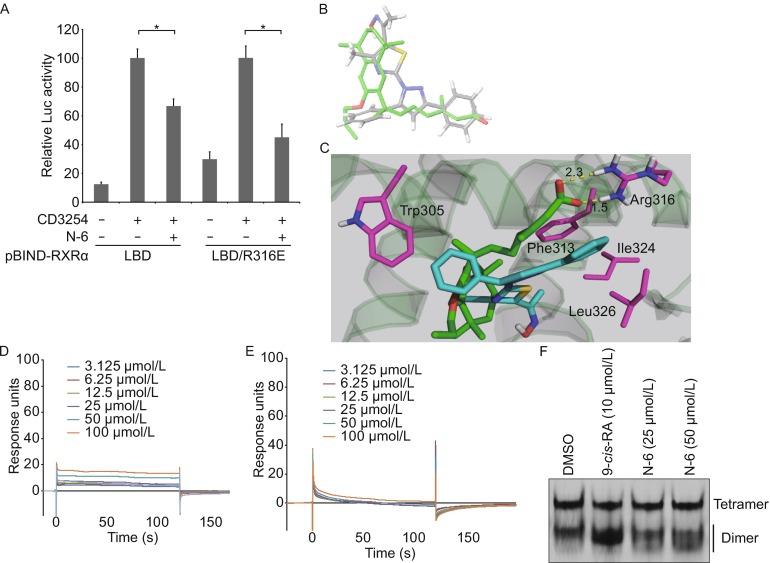
Arg316 is not required for N-6 binding to RXRα. (A) MCF-7 cells cotransfected with pG5-Gaussia-Dura reporter vector and pBIND-RXRα-LBD or pBIND-RXRα-LBD/R316E expression vectors were treated with or without N-6 (10 μmol/L) in the presence or absence of CD3254 (10−7 mol/L) for 18 h. Reporter activities were measured and normalized. Data shown are mean ± SD (*P < 0.05). (B) Comparison of the docked conformation of N-6 (gray) with the crystal structure of LG100754 (green). (C) N-6 was docked into the LBP of the co-crystal structure of LG100754 and RXRα-LBD (PDB 3A9E). Salt bridges are shown as dotted yellow lines, and residues interacting with N-6 are shown in magenta. (D–E) Gradient concentrations of N-6 were injected through flow cells immobilized with RXRα-LBD/Trp305Ala (D) and RXRα-LBD/Phe313Ala (E), respectively. The kinetic profiles are shown and the dissociation constants (K d) of the N-6/RXRα-LBD complex were calculated to be 1.0 × 10−3 mol/L (D) and more than 1.0 × 10−2 mol/L (E). (F) RXRα-LBD proteins (2 mg/mL) was incubated with DMSO, 10 μmol/L 9-cis-RA, 25 μmol/L N-6 or 50 μmol/L N-6 for 3 h, and proteins were separated by 8% nondenaturing PAGE followed by Commassie Blue staining
