Figure 6.
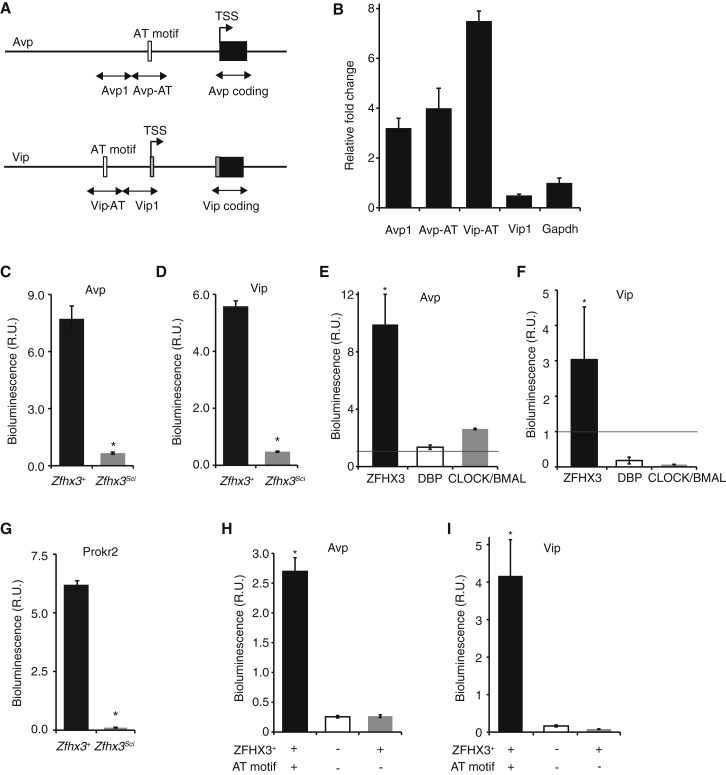
ZFHX3SCI Interacts with and Differentially Activates the AT Motif in Avp and Vip Promoters
Quantitative ChIP of Zfhx3+/+ SCN tissue samples using ZFHX3 antiserum.
(A) Primer pairs were designed to span the AT motif (Avp-AT/Vip-AT) and an adjacent region (Avp-1/Vip-1) upstream of the TSS (primers in the coding region of each gene were used to normalize each reaction).
(B) Zfhx3 binds to the promoter around the AT motif of both Avp and Vip compared to the control gene, Gapdh. Data are shown as fold change normalized for input relative to the corresponding coding region of each gene and are taken at ZT3 (n = 3).
(C and D) Overexpression of ZFHX3 with the Sci mutation (ZFHX3Sci) was ineffective in activation of luciferase driven by AT-motif-containing promoters of (C) Avp and (D) Vip in HEK293 cells (p < 0.05, t test). R.U., relative units.
(E and F) Zfhx3+ activation (black bars) of the (E) Avp and (F) Vip promoters was more than three times that seen for either DBP (white bars) or CLOCK/BMAL (gray bars) (p < 0.05, t test).
(G) Overexpression ZFHX3Sci was also ineffective in activation of the Prokr2 promoter Prokr2 in HEK293 cells (p < 0.05, t test).
(H and I) Mutation of the three most conserved residues (positions 6, 8, and 10) in the AT motifs of both the (H) Avp and (I) Vip promoter constructs (gray bars) resulted in significantly decreased levels of ZFHX3+ activation compared to those with AT motifs intact (black bars) (p < 0.05, t test). White bars represent the activation of the mutated promoter constructs alone.
Error bars indicate SEM. ∗p < 0.05, t test.
See also Figure S5.