Fig. 1.
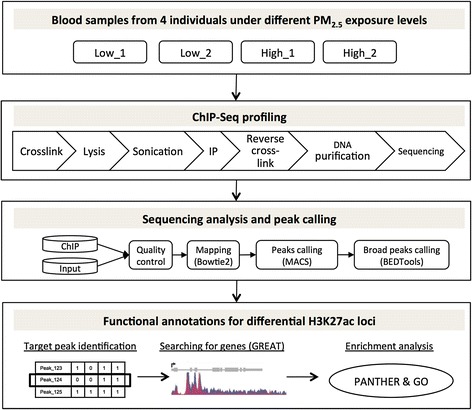
Overview of the study design and analysis workflow. Nuclei were extracted from blood samples of four individuals under low or high exposure of outdoor PM2.5. Standard protocol was used for ChIP-Seq experiments. Bioinformatics software (in brackets) and in-house scripts were used to analyze the sequencing data. The functions of differential H3K27ac loci were evaluated using public databases. ChIP: chromatin immunoprecipitation; IP: immunoprecipitation; GO: gene ontology
