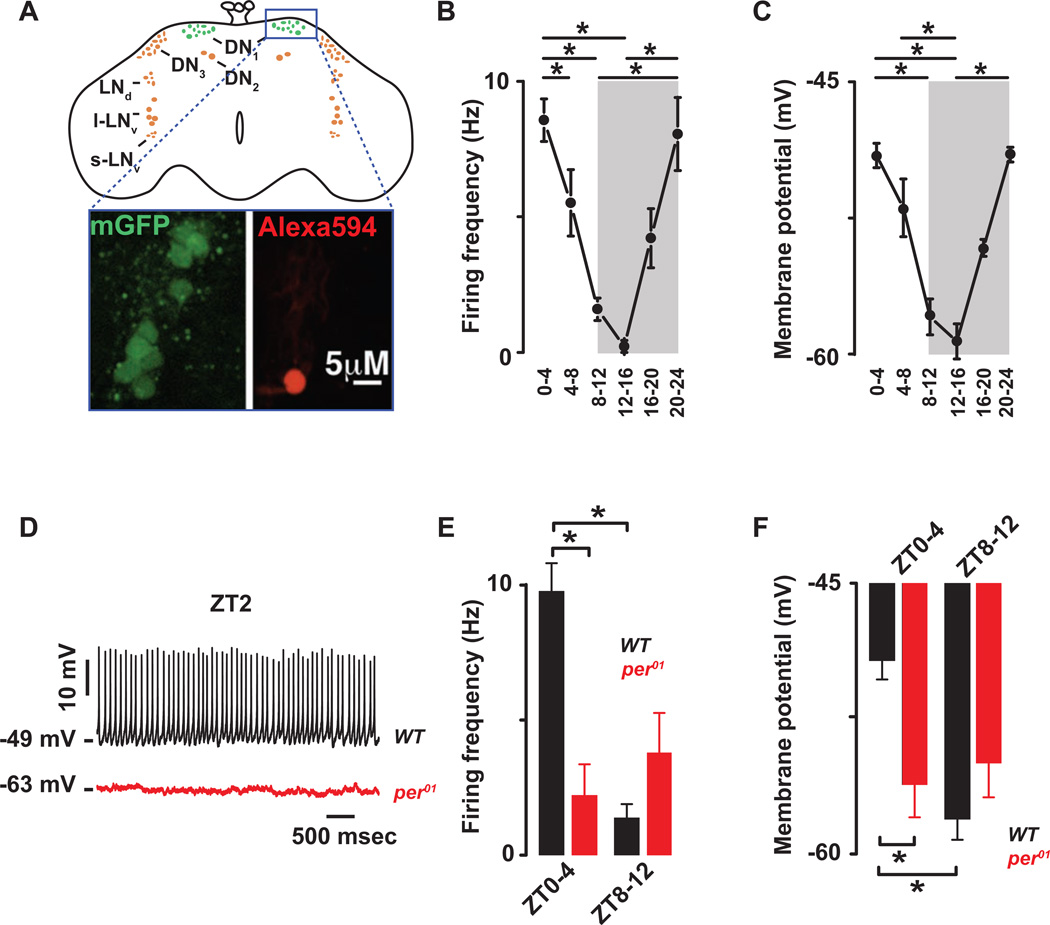
Figure 1. The cellular excitability of the Drosophila DN1p circadian pacemaker neurons is clock controlled.
(A) Schematic and image of the Drosophila brain indicating the location of the DN1ps and other clock neurons. Representative images of the GFP-expressing DN1ps in the intact Drosophila brain are shown below. The DN1ps were labeled by using the Clk4.1M–G4 driving the expression of U-CD8-GFP. Whole-cell access to GFP labeled neurons was confirmed following diffusion of Alexa Fluor 594 biocytin included in intracellular recording solution. All recorded WT neurons are plotted against time of day (in 4 hours bins) to show daily rhythms of firing frequency (B) and membrane potential (C). Grey areas represent the dark phase of the LD cycle. Asterisks indicate statistical significance (p<0.05) from a one-way ANOVA, Tukey’s post-hoc test. (D) Representative current clamp recordings at Zeitgeber Time-2 (ZT2) showing that the per01 DN1ps neurons (red) are hyperpolarized and silent compared to WT DN1p neurons (black). Histogram showing the decrease in firing frequency (E) and membrane potential (F) and lack of daily rhythm in per01 (red, 2.2±1.1Hz, −56±2 mV, n=15 at ZT0-4 and 3.9±1.5Hz, −55±1.9 mV, n=10 at ZT8–12, p>0.41) when compared to WT (black) DN1p neurons. Results are expressed as mean±SEM. Asterisks indicate statistical significance (p<0.05) from t-test performed in WT at ZT0-4 vs ZT8–12. See also Figures. S.1–2 and Tables S.1–2.