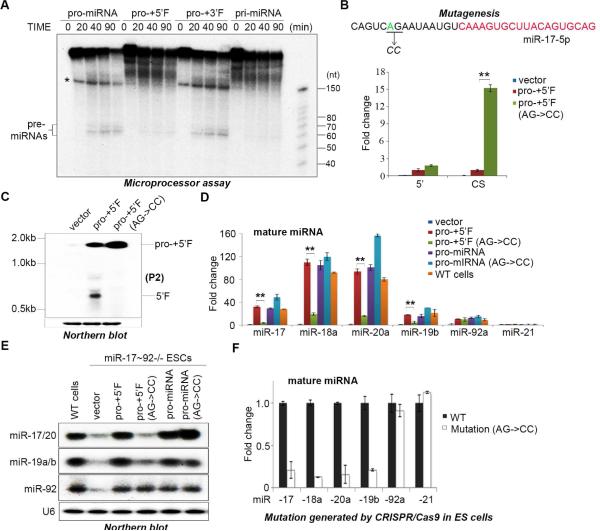
Figure 2. Cleavage of pri-miR-17~92 to pro-miRNA is a key step in miRNA maturation.
(A) Microprocessor cleavage assays performed using the indicated substrates. Asterisk denotes a truncated or non-specific RNA. (B) q.RT-PCR analysis of the relative expression of regions of miR-17~92 expressed from the indicated plasmids. Primers amplifying the 5’ upstream sequence (5’) and primers spanning the cleavage site (CS) were used to detect pri-miR-17~92 in transfected miR-17~92−/− ESCs. Data normalized to ACTIN and represented as mean +/− SEM. **p < 0.01, Student's t test. (C) Northern blot analysis of the RNA samples in (B) using a probe that detects the 5’ upstream region of pri-miR-17~92 (P2). (D) miR-17~92−/− ESCs were transfected with the indicated rescue plasmids and mature miRNAs measured by q.RT-PCR. Data normalized to snoR142 and represented as mean +/− SEM. **p < 0.01, Student's t test. (E) Northern blot analysis of the RNAs from (D) using probes to detect the indicated miRNAs. (F) q.RT-PCR analysis of the indicated endogenous mature miRNAs expression in ESCs engineered with a mutation in the cleavage site at the endogenous pri-miR-17~92 locus. Data are normalized to snoR142 and represented as mean +/− SEM. See also Supplementary Figure 1.