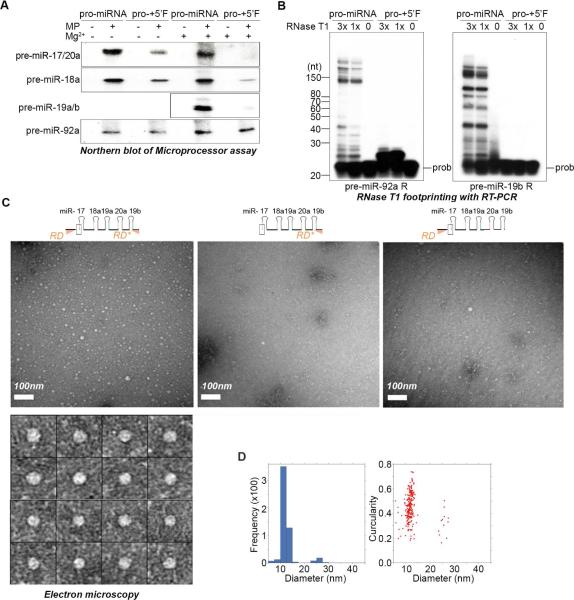
Figure 4. Pri-miR-17~92 adopts an RNA conformation that inhibits Microprocessor.
(A) Microprocessor cleavage assays performed with (+) or without (−) RNA annealing in the presence of MgCl2. Reaction products were loaded onto multiple gels and analyzed by Northern blot. (B) RNase T1 accessibility assays performed using the indicated RNA and analyzed by reverse transcriptase primer extension using the indicated 5’-end labeled primers. (C) Negative-stain micrographs of indicated RNAs in the presence of MgCl2. Specimens were prepared in uranyl acetate. Lower panel shows representative images of RD-Pro-RD* particles. (D) 2D distribution of RD-Pro-RD* particles based on their diameter and circularities.