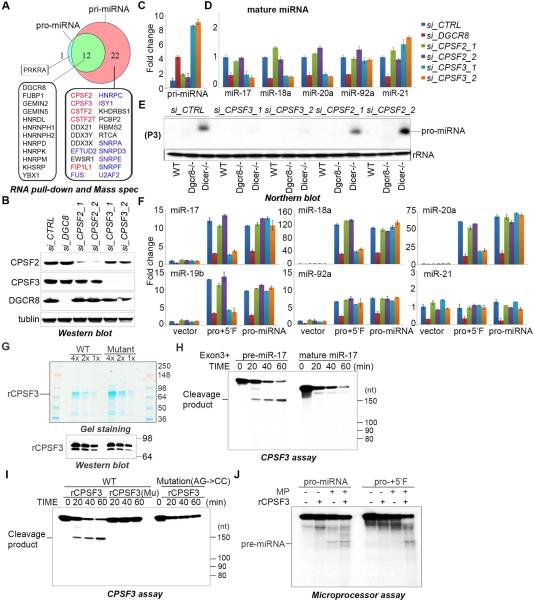
Figure 5. CPSF3 endonuclease is required for pro-miRNA biogenesis and mature miRNA expression.
(A) Summary of mass spec analysis of RNA-affinity purifications. Factors known to be involved in pre-mRNA 3’ cleavage and polyadenylation are highlighted in red and proteins involved in splicing are listed in blue. (B) Western blot of lysates prepared from ESCs transfected with the siRNAs and analyzed using the indicated antibodies. (C) q.RT-PCR analysis of pri-miRNA expression in cells with indicated siRNA knockdown. Data normalized to ACTIN and represented as mean +/− SEM. (D) q.RT-PCR analysis of the indicated endogenous miRNAs in ESCs transfected with the siRNAs shown. Data normalized to snoR142 and represented as mean +/− SEM. (E) Northern blot performed on total RNA, from wt, Dgcr8−/−, and Dicer−/− ESCs transfected with the indicated siRNAs. A probe was used to detect pro-miRNA as indicated on the right (P3). (F) q.RT-PCR analysis of the relative expression the indicated miRNAs expressed from the indicated miR-17~92 rescue plasmids co-transfected with the indicated siRNAs. Data normalized to snoR142 and represented as mean +/− SEM. (G) Coomassie blue stained SDS-PAGE gel (top) and αCPSF3 western analysis (bottom) of recombinant His-CPSF3 purified from E.coli. Wild-type (WT) and a catalytic mutant CPSF3 (D75K/H76A) were produced. (H, I) CPSF cleavage assays using the indicated in vitro transcribed RNA substrate and His-CPSF3 (WT or Mutant). (J) Microprocessor cleavage assay with the indicated RNA substrate and with addition of His-CPSF3 where indicated. See also Supplementary Figure 3.