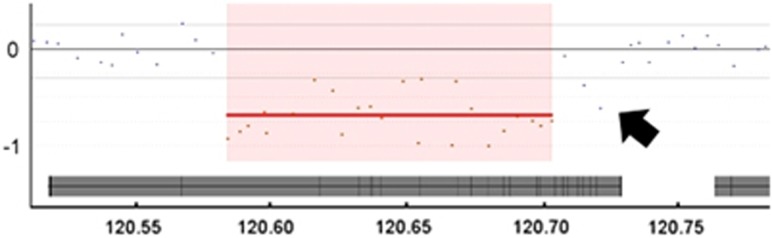
Figure 1.
Comparative genomic hybridisation (CGH) array analysis. CGH array testing mapped the approximate position of the deletion, at coordinates 120.58–120.70 Mb, within chromosome 2. The 400K array data shown here is for Twin A. The central grey bar indicates gene PTPN4. The x axis shows the hg19 coordinates. This image is derived from the CGH Fusion software: InfoQuant Ltd (London, UK). Note that the segmentation algorithm used here may have underestimated the distal extent of the deletion (see arrow). Within the ISCN terminology, the deletion was reported as extending from 120,584,760 to 120,726,563.