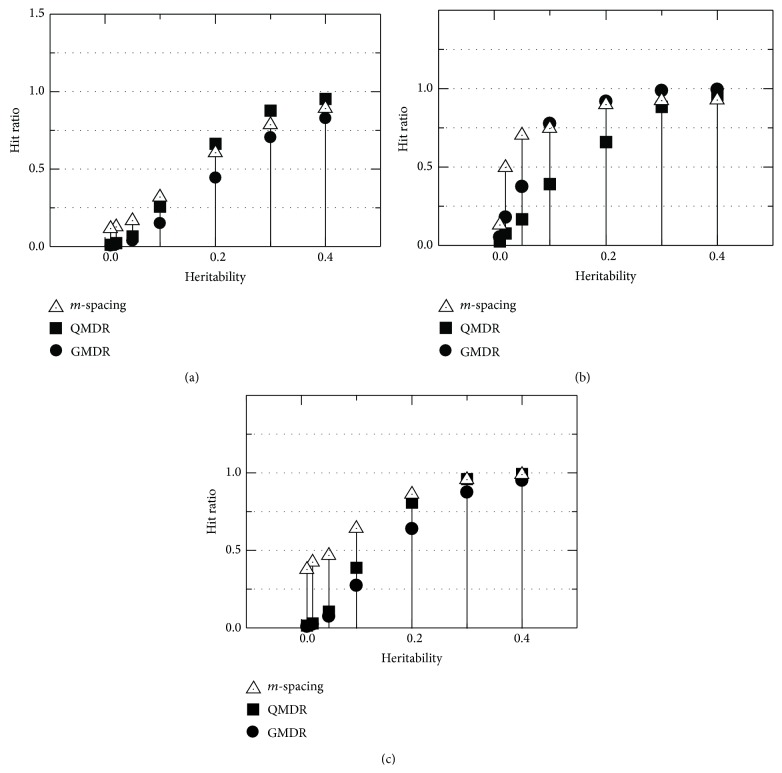
Figure 5.
Comparison of the hit ratios or the detection probabilities among the proposed m-spacing method, QMDR, and GMDR. Genomic datasets were generated based on 70 different penetrance functions [21], which were, in turn, classified into 7 distinct values of heritability. For each model, the phenotype values are simulated with normal (a), gamma (b), and mixed (c) distributions. High- and low-risk groups in a quantitative trait overlapped with 9 different combinations of the standard deviations. Considering all of the above, 100 data files were generated for each case, adding up to 9,000 simulated files being examined for each point in the plot.