Abstract
The Cfr RNA methyltransferase causes multiple resistances to peptidyl transferase inhibitors by methylation of A2503 23S rRNA. Many cfr-like gene sequences in the databases code for unknown functions. This study confirms that a Cfr-like protein from a Peptoclostridium difficile (formerly Clostridium difficile) strain does function as a Cfr protein. The enzyme is expressed in Escherichia coli and shows elevated MICs for five classes of antibiotics. A primer extension stop indicates a modification at A2503 in 23S rRNA.
TEXT
The cfr gene was initially found on a plasmid in coagulase-negative staphylococci of animal origin (1). A very recent paper titled “Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr” presented MIC data on linezolid susceptibility that indicate the presence of a cfr-like gene that may provide antibiotic resistance by the same mechanism as Cfr (2). As pointed out by Schwarz and Wang (3), such a statement requires proof that the gene actually expresses an enzyme with that function. We therefore introduced a gene coding for the same enzyme, herein named clcD, into a plasmid and transformed the plasmid into Escherichia coli to analyze for a Cfr-like function. The gene sequence was optimized to E. coli codon usage, synthesized, and inserted as described in reference 4. After transformation to E. coli AS19 (5) and E. coli JW2505-1 (ΔRlmN) (6), MICs and modification at A2503 23S rRNA were investigated as previously presented in a similar study with cfr-like genes from Bacillales (4).
The MICs in Table 1 were determined as described in reference 4 and showed that IPTG (isopropyl-β-d-thiogalactopyranoside) induction of expression of the clcD gene can provide resistance to representatives of five of the six antibiotic groups previously shown to be affected by Cfr (7, 8). The resistance was not as strong as the one seen with Cfr in this setup, which was similar to that with the original cfr gene in pBglII (4), but the resistance was comparable to some of those seen from Bacillales Cfr-like proteins (4). To further establish the function of ClcD as similar to that of Cfr, which causes 8-methyladenosine (m8A) methylation at A2503 in 23S rRNA (9), we examined the A2503 position by primer extension. The pClcD plasmid was introduced into E. coli JW2505-1 (ΔRlmN) to remove interference from the weak primer extension stop known to result from RlmN m2A2503 methylation (9). After induction, total RNA was isolated and hybridized with a primer specific for a region downstream of A2503, followed by extension by reverse transcriptase and gel electrophoresis, all performed as described previously (4). An m8A modification will cause a band on the gel due to pausing. The primer extension data clearly showed a primer extension stop at A2503 dependent on the presence of ClcD and at the same position as the stop caused by Cfr (Fig. 1). We also observed a reduction of the Cm2498 stop as seen with Cfr (7). The combined information of a modification at A2503 and increased MICs from five different antibiotics in the presence of ClcD shows that the strain investigated by Marin et al. (2) contains a gene that codes for a protein with a function similar to that of Cfr. The antibiotic resistance seen in Peptoclostridium difficile strain 11140508 can thus be considered the result of expression of this gene (2).
TABLE 1.
MICs of the hypersensitive E. coli AS19 with the plasmid-coded ClcD enzyme versus the strain alone, the strain with the parent plasmid, and strains with plasmid-coded Cfrhis
| Plasmid | cfr/cfr-like genea | MICb (μg/ml) for: |
||||
|---|---|---|---|---|---|---|
| Florfenicol | Clindamycin | Linezolid | Tiamulin | Synercid | ||
| None | − | 1 | 32 | 8 | 0.25 | 1 |
| pLJ102 | − | 1 | 16 | 8 | 0.25 | 1 |
| pCfrhis | + | 16 | >128 | 128 | 16 | 16 |
| pClcD | + | 8 | >128 | 32 | 4 | 4 |
The cfr/cfr-like genes are expressed only if induced by IPTG. To see the phenotype from expression of these genes (the antibiotic resistance), IPTG was added prior to growth in the presence of antibiotics, for the MIC determinations. +, presence of a plasmid with a cfr or cfr-like gene; −, absence of a cfr or cfr-like gene.
MICs determined as described in reference 4.
FIG 1.
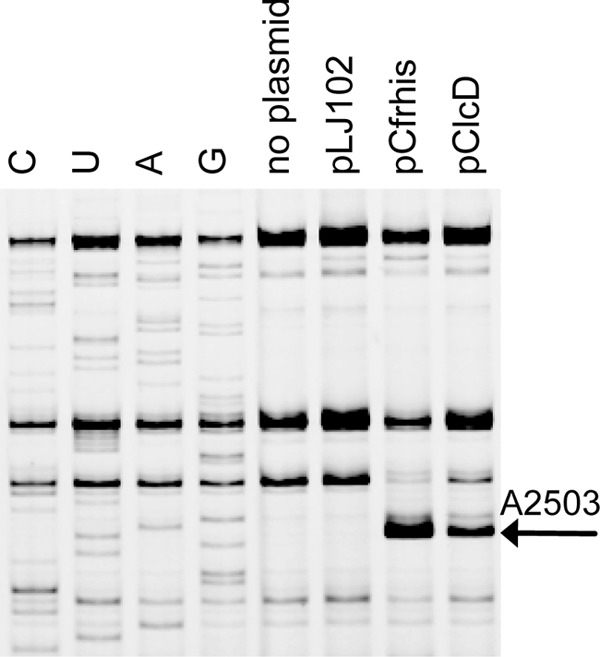
Primer extension analysis of reverse transcriptase stops on 23S rRNA from E. coli JW2501-1 harboring the plasmid expressing the ClcD protein. The region shown is limited to the nucleotides flanking A2503. Lanes 1 to 4 (C, U, A, and G), dideoxy sequencing reactions. Lanes 5 to 8, primer extension reactions on total RNA from cells harboring the indicated plasmids. Reverse transcriptase stops 1 nucleotide before the corresponding nucleotide in the sequencing lanes. Arrow, stop mediated by modification from Cfr or Cfr-like proteins.
A common way to find a function for an uninvestigated gene and its coded protein is to perform a BLAST search to look for similar proteins. In the case of Cfr, the interpretation of such searches is not straightforward because few of the proteins have been investigated and some annotations are flawed. Sequences of the relatively similar RlmN enzymes are included, and some of the proteins are misannotated (Cfr-like proteins as RlmN and vice versa). We previously investigated sequence differences to distinguish between Cfr-like proteins and RlmN (10). Unfortunately, there is not yet enough information to tell which Cfr-like proteins function as Cfr (making m8A2503 and causing resistance) and which do not. We have some preliminary and unpublished data indicating that not all Cfr-like proteins function as Cfr. The Cfr-like protein in P. difficile (formerly Clostridium difficile [11]) is hosted by a transposon, and the same gene is found in other strains (2). Figure 2 shows an alignment of a small selection of Cfr-like proteins from a BLAST search compared to Cfr (75% identity to ClcD) to illustrate the task of assigning function to these proteins. BLAST showed that ∼10 P. difficile and Enterococcus faecalis strains contain a clcD gene (≥99% identity) (Fig. 2A). More than 40 Bacillales also contain genes for similar proteins (84% to 87% identity to ClcD, 57% to 77% identity to Cfr), and 3 of these were shown to function as Cfr (4). In contrast, it has not yet been possible to establish that the Cfr-like sequences represented in Fig. 2D (>40 sequences, 56% to 57% identity to ClcD and Cfr and found in Clostridium sporogenes and Clostridium botulinum) function as Cfr (our unpublished data). So far, there is no clear correlation with a precise minimum percentage identity to tell whether a protein has the Cfr function.
FIG 2.
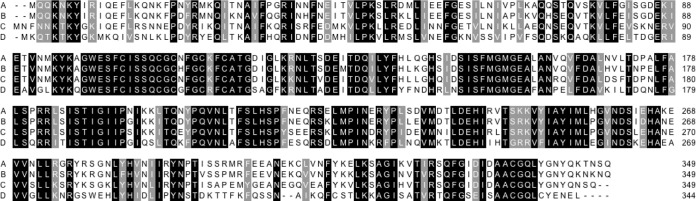
Protein alignments of ClcD represented by KM359438.1, GI:723115824, Peptoclostridium difficile strain 11140508 (A), of Cfr-like proteins from Bacillales represented by WP_012116915.1, GI:501065687, Bacillus amyloliquefaciens (B), of Cfr from Staphylococcus sciuri represented by NP_899167.1, GI:34328031 (C), and of Cfr-like proteins from Clostridium sporogenes and Clostridium botulinum represented by WP_012100648.1, GI:501048958 (D). Black shading, 100% identity for the 4 sequences; gray shading, similarity of 3 out of 4 using the groups ILV, FWY, KRH, DE, GAS, P, C, and TNQM. The alignment was made using the Sequence Manipulation Suite (http://www.bioinformatics.org/sms/).
Cfr is becoming a real threat for combating some pathogenic bacteria due to its widespread occurrence (12), and Cfr-like proteins may also be found to provide antibiotic resistance. It is thus important to establish the dissemination and function of Cfr-like proteins to know what to look for and be able to take appropriate precautions.
ACKNOWLEDGMENT
We thank the Danish National Research Foundation for financial support (grant 12-125943).
REFERENCES
- 1.Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44:2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin M, Martin A, Alcala L, Cercenado E, Iglesias C, Reigadas E, Bouza E. 2015. Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob Agents Chemother 59:586–589. doi: 10.1128/AAC.04082-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz S, Wang Y. 2015. Nomenclature and functionality of the so-called cfr gene from Clostridium difficile. Antimicrob Agents Chemother 59:2476–2477. doi: 10.1128/AAC.04893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen LH, Planellas MH, Long KS, Vester B. 2012. The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob Agents Chemother 56:3563–3567. doi: 10.1128/AAC.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekiguchi M, Iida S. 1967. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci U S A 58:2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 8.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkinson GC, Hansen LH, Tenson T, Rasmussen A, Kirpekar F, Vester B. 2013. Distinction between the Cfr methyltransferase conferring antibiotic resistance and the housekeeping RlmN methyltransferase. Antimicrob Agents Chemother 57:4019–4026. doi: 10.1128/AAC.00448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]


