Figure 8.
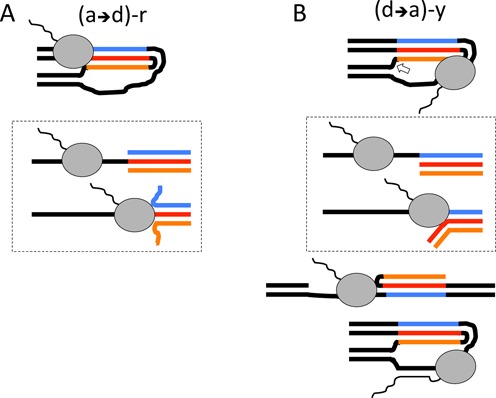
Possible mechanisms of transcription blockage by stable H-DNA analog in various orientations. (A) The simplest mechanism of blockage is expected for (a→d)-r configuration, in which RNA polymerase (RNAP) (gray oval) is bumping into the triplex and is unable to unwind it. In this case, the template strand (red) is involved in Watson–Crick and Hoogsteen interactions and RNA polymerase has to disrupt the triplex as a whole (see the scheme in the dashed line-border box below). At the same time, two displaced strands are not pairing with each other resulting in the high entropic barrier for triplex re-formation. (B) The mechanism of blockage in the case of (d→a)-y configuration is more complex: on one hand, the energetic barrier for the triplex dismantling is lower, since only Watson–Crick interactions have to be disrupted to make the template available; on the other hand Hoogsteen interactions at the displaced strands could persist, decreasing the entropic barrier for triplex reformation (see the scheme in the dashed line-border box below). Altogether, it is easier for an elongating RNAP to unwind the triplex in (d→a)-y than in (a→d)-r construct (in Figure 8, A), but the probability that it would be ‘pushed back’ by triplex re-formation is greater in (d→a)-y construct. In the (d→a)-y configuration, RNAP could be sequestered by sterical constraints even before it started to unwind the triplex. This sequestration is gone if the H-DNA-like structure is cut at its triplex-duplex junction (bottom). Finally, hybridization between the nascent RNA and the template strand (bottom) could also contribute to the blockage in the (d→a)-y configuration.
