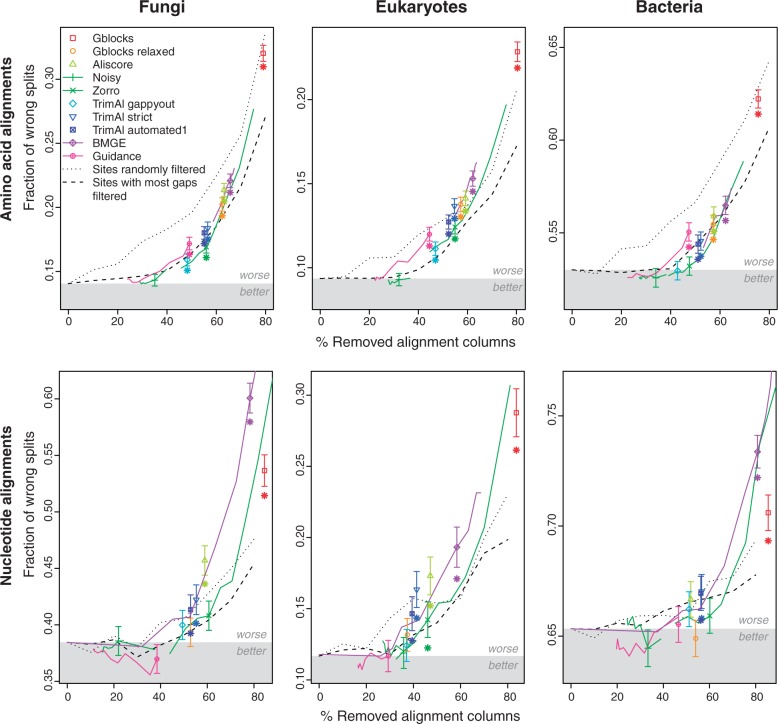
Figure 2.
Alignment filtering generally yields poorer phylogenetic trees. Depicted here are results with the enriched species tree discordance test on amino acid (top) and nucleotide (bottom) alignments from three taxonomic ranges. The measure of error is the average RF distance between the reference trees and trees reconstructed from Prank + F alignments filtered by the various approaches. Trees were reconstructed using PhyML. Filtered alignments improving over unfiltered alignment fall in the gray region. The two dotted lines correspond to results obtained with two simplistic filtering methods (see main text). Points correspond to default parameters. Colored lines are linear interpolations between additional points obtained with non-default parameters (not available for all methods). Error bars indicate the standard error of the mean. If a filtering method with default parameters yields significantly different (two-sided Wilcoxon test, ) results from unfiltered alignments, a star is displayed below the corresponding point. Note that no multiple testing correction were applied.