Fig. 4.
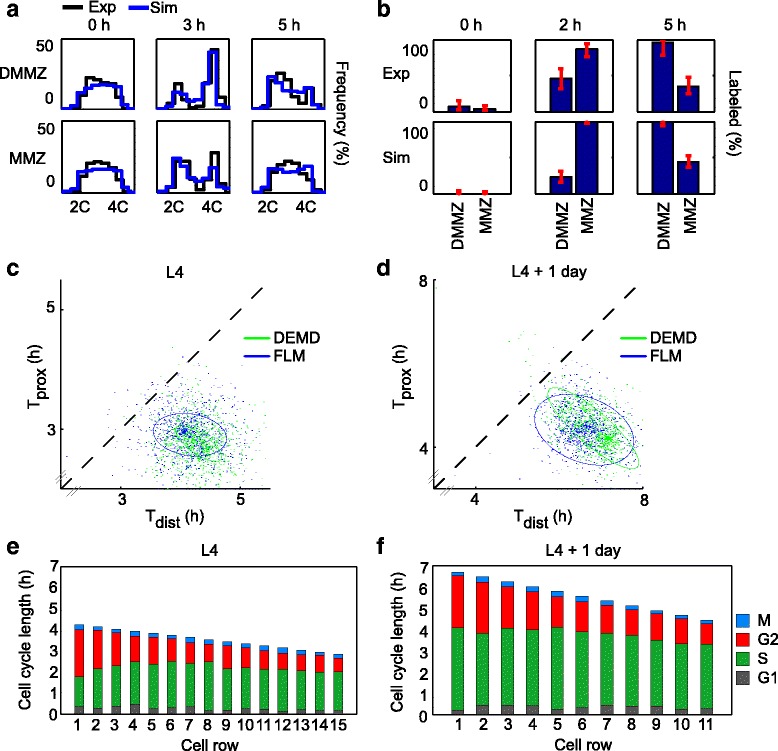
Quantitative cell cycle models that allow for a cell cycle gradient across the MZ provide a good fit to experimental data, and show ~1.5-fold slower cycling of stem cells. a DNA content histograms of EdU-positive cells derived from best-fit simulations of cell cycling to L4 + 1 day experimental data (black) overlaid with the same experimental data (blue), at 0 h, 3 h, and 5 h (full overlay shown in Additional file 2: Figure S1). Experimental data were derived from a total of n = 157 gonadal arms. b Fractions of EdU-labeled mitoses derived from L4 + 1 day experimental data (“Exp” row) or from best-fit simulations (“Sim” row; full overlay shown in Additional file 2: Figure S1). c, d Best-fit cell cycle parameters show faster cell cycling at the proximal end of the MMZ (y-axis) than at the distal DMMZ (x-axis) both at L4 (c) and L4 + 1 day (d), and both when fitting DNA content histograms (DEMD; green) or fractions of labeled mitoses (FLM; blue). Each dot on the graph corresponds to a bootstrap sample; ellipses contain 95 % of bootstrap samples and are located off the diagonal, which corresponds to equal cell cycle speeds across the distal–proximal axis. Jitter was added to bootstrap samples to aid visualization (see Additional file 2: Figure S2 for display without jitter). e, f Distal cells have longer G2 than proximal cells. Stacked bars show the length of each cell cycle phase along the distal–proximal axis, as computed using best-fit parameters. Note that absolute cell cycle lengths cannot be directly derived from Fig. 3b
